8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 2024
|
|
8R6F
 
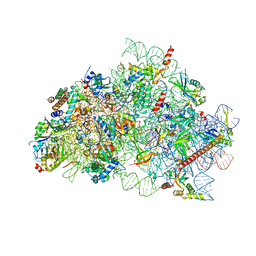 | | CryoEM structure of wheat 40S ribosomal subunit, body domain | | Descriptor: | 30S ribosomal protein S11, chloroplastic, 30S ribosomal protein S4, ... | | Authors: | Kravchenko, O.V, Baymukhametov, T.N, Afonina, Z.A, Vasilenko, K.S. | | Deposit date: | 2023-11-22 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | High-Resolution Structure and Internal Mobility of a Plant 40S Ribosomal Subunit.
Int J Mol Sci, 24, 2023
|
|
8R67
 
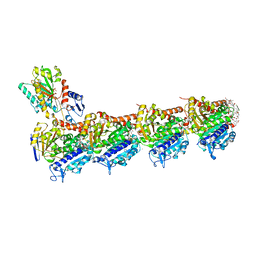 | | tubulin-cryptophycin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(3~{S},10~{R},13~{E},16~{S})-10-[(3-chloranyl-4-methoxy-phenyl)methyl]-6,6-dimethyl-2,5,9,12-tetrakis(oxidanylidene)-16-[(1~{S})-1-[(2~{R},3~{R})-3-phenyloxiran-2-yl]ethyl]-1,4-dioxa-8,11-diazacyclohexadec-13-en-3-yl]ethanoic acid, CALCIUM ION, ... | | Authors: | Abel, A.C, Muehlethaler, T, Dessin, C, Steinmetz, M.O, Sewald, N, Prota, A.E. | | Deposit date: | 2023-11-21 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bridging the maytansine and vinca sites: Cryptophycins target beta-tubulin's T5-loop.
J.Biol.Chem., 300, 2024
|
|
8X6P
 
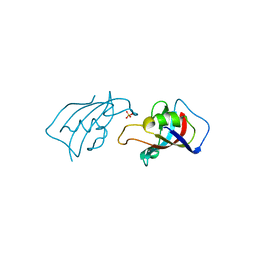 | | Isomerase Protein | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | Authors: | Guven, O, Demirci, H. | | Deposit date: | 2023-11-21 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Isomerase Protein
To Be Published
|
|
8X6R
 
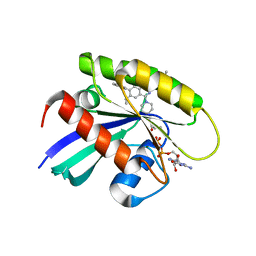 | | KRasG12C in complex with inhibitor | | Descriptor: | 1-[7-[6-ethenyl-8-ethoxy-7-(5-methyl-1~{H}-indazol-4-yl)-2-(1-methylpiperidin-4-yl)oxy-quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of ASP6918, a KRAS G12C inhibitor: Synthesis and structure-activity relationships of 1-{2,7-diazaspiro[3.5]non-2-yl}prop-2-en-1-one derivatives as covalent inhibitors with good potency and oral activity for the treatment of solid tumors.
Bioorg.Med.Chem., 98, 2023
|
|
8X6M
 
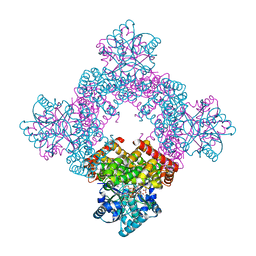 | | Crystal Structure of Glycerol Dehydrogenase in the Presence of NAD+ and Glycerol | | Descriptor: | GLYCEROL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Kang, J.Y, Jin, M, Yang, J, Kim, H, Noh, C, Eom, S.H. | | Deposit date: | 2023-11-21 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the octamerization of glycerol dehydrogenase.
Plos One, 19, 2024
|
|
8X6G
 
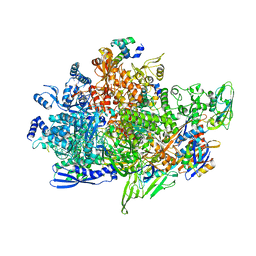 | | Cryo-EM structure of Staphylococcus aureus sigB-dependent RNAP-promoter open complex | | Descriptor: | DNA (70-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Nat Commun, 15, 2024
|
|
8X6F
 
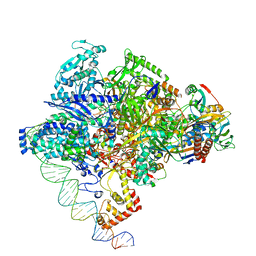 | | Cryo-EM structure of Staphylococcus aureus sigA-dependent RNAP-promoter open complex | | Descriptor: | DNA (71-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Nat Commun, 15, 2024
|
|
8R65
 
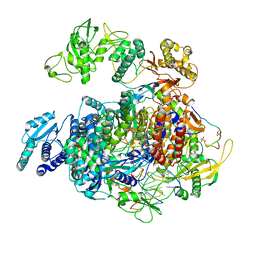 | | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide with ordered PB2 C-terminal domains | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide with ordered PB2 C-terminal domains
To Be Published
|
|
8R62
 
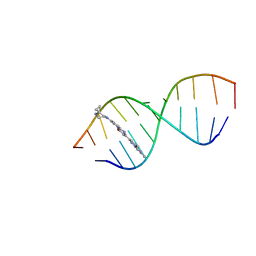 | | Solution structure of Risdiplam bound to the RNA duplex formed upon 5'-splice site recognition | | Descriptor: | 7-(4,7-diazaspiro[2.5]octan-7-yl)-2-(2,8-dimethylimidazo[1,2-b]pyridazin-6-yl)-1~{H}-pyrido[1,2-a]pyrimidin-4-one, RNA (5'-R(*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Campagne, S. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
8V1H
 
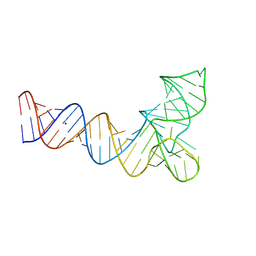 | |
8V1I
 
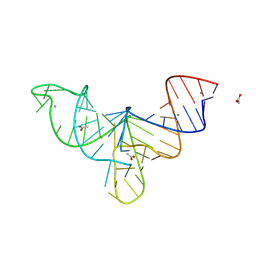 | | Crystal structure of human mascRNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Skeparnias, I, Zhang, J. | | Deposit date: | 2023-11-20 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of MALAT1 RNA maturation and mascRNA biogenesis
Nat.Struct.Mol.Biol., 2024
|
|
8R63
 
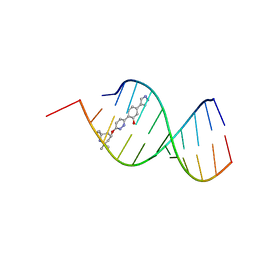 | | Solution structure of branaplam bound to the RNA duplex formed upon 5'-splice site recognition | | Descriptor: | 5-(1~{H}-pyrazol-4-yl)-2-[6-(2,2,6,6-tetramethylpiperidin-4-yl)oxypyridazin-3-yl]phenol, RNA (5'-R(*AP*UP*AP*CP*(PSU)P*(PSU)P*AP*CP*CP*UP*G)-3'), RNA (5'-R(P*GP*GP*AP*GP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Malard, F, Campagne, S. | | Deposit date: | 2023-11-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The diversity of splicing modifiers acting on A-1 bulged 5'-splice sites reveals rules for rational drug design.
Nucleic Acids Res., 52, 2024
|
|
8V1L
 
 | |
8R60
 
 | | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-19 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide
To Be Published
|
|
8X5V
 
 | | BlCas9-sgRNA-target DNA complex | | Descriptor: | 1,2-ETHANEDIOL, BlCas9, CHLORIDE ION, ... | | Authors: | Nakane, T, Nakagawa, R, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2023-11-19 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and engineering of Brevibacillus laterosporus Cas9.
Commun Biol, 7, 2024
|
|
8V15
 
 | | Human SIRT3 bound to p53-AMC peptide, Carba-NAD, and Honokiol | | Descriptor: | (1P)-3',5-di(prop-2-en-1-yl)[1,1'-biphenyl]-2,4'-diol, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, ... | | Authors: | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | Deposit date: | 2023-11-19 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8R5O
 
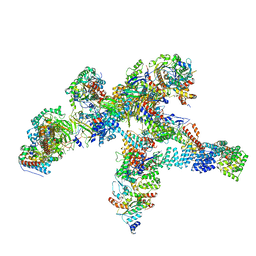 | | Plastid-encoded RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8V08
 
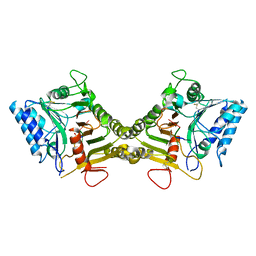 | |
8V06
 
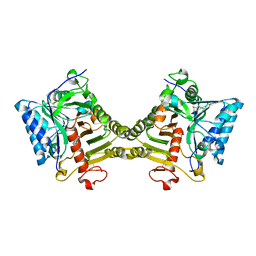 | | Crystal structure of mouse PLD3 co-crystallized with 5'Pi-ssDNA for 9 days | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4.
Structure, 32, 2024
|
|
8V05
 
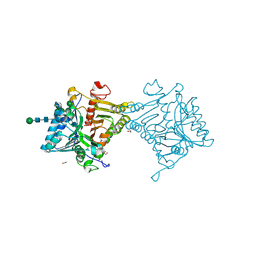 | | Crystal structure of mouse PLD3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4.
Structure, 32, 2024
|
|
8V07
 
 | | Crystal structure of mouse PLD3 co-crystallized with 5'Pi-ssDNA for 30 days | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4.
Structure, 32, 2024
|
|
8X5L
 
 | | The Crystal Structure of PRKACA from Biortus. | | Descriptor: | (2S)-2-(4-chlorophenyl)-2-hydroxy-2-[4-(1H-pyrazol-4-yl)phenyl]ethanaminium, SODIUM ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Wang, F, Cheng, W, Lv, Z, Lin, D, Pan, W. | | Deposit date: | 2023-11-17 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of PRKACA from Biortus.
To Be Published
|
|
8X5D
 
 | |
8R5V
 
 | | Crystal structure of bovine pancreatic ribonuclease A in complex with [Sp-PS]-mU-dT dinucleotide | | Descriptor: | 1-[(2~{R},4~{S},5~{R})-5-[[[(2~{R},6~{S})-2-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-6-(hydroxymethyl)morpholin-4-yl]-oxidanyl-phosphinothioyl]oxymethyl]-4-oxidanyl-oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione, DICHLORO-ACETIC ACID, Ribonuclease pancreatic | | Authors: | Dolot, R, Jastrzebska, K, Antonczyk, P. | | Deposit date: | 2023-11-17 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of P-stereodefined morpholino dinucleoside-3',6'-thiophosphoramidates using unexpected activator.
To Be Published
|
|
