1AN5
 
 | | E. COLI THYMIDYLATE SYNTHASE IN COMPLEX WITH CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
4ORR
 
 | | Threedimensional structure of the C65A mutant of Human lipocalin-type Prostaglandin D Synthase olo-form | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Prostaglandin-H2 D-isomerase, SULFATE ION | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OS0
 
 | | Three-dimensional structure of the C65A-W54F double mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8QMI
 
 | | Crystal structure of RNA G2C4 repeats - native model | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Blaszczyk, L, Kiliszek, A. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
5N75
 
 | | 14-3-3 sigma in complex with TAZ pS89 peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-19 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
5N5R
 
 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV1 | | Descriptor: | 14-3-3 protein sigma, 2-azanyl-~{N}-(2,6-dimethylphenyl)-~{N}-propan-2-yl-ethanamide, CHLORIDE ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
2GL8
 
 | | Human Retinoic acid receptor RXR-gamma ligand-binding domain | | Descriptor: | Retinoic acid receptor RXR-gamma | | Authors: | Min, J.R, Schuetz, A, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-04 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Human Retinoic acid receptor RXR-gamma ligand-binding domain.
To be Published
|
|
6ZFL
 
 | | High resolution structure of VEGF-A 12:107 crystallized in tetragonal form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Vascular endothelial growth factor A | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
8QMH
 
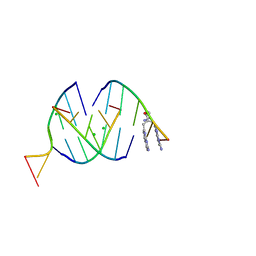 | | Crystal structure of RNA G2C4 repeats in complex with small synthetic molecule ANP77 | | Descriptor: | 3-(7-azanyl-1,8-naphthyridin-2-yl)-2-[(7-azanyl-1,8-naphthyridin-2-yl)methyl]-~{N}-(3-azanylpropyl)propanamide, CHLORIDE ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Kiliszek, A, Ryczek, M, Blaszczyk, L. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
8EAQ
 
 | | Structure of the full-length IP3R1 channel determined at high Ca2+ | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 1, ... | | Authors: | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
8EAR
 
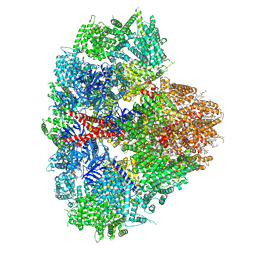 | | Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
20GS
 
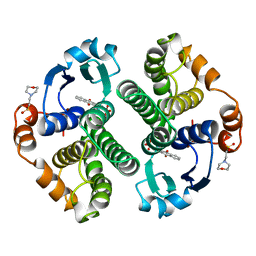 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH CIBACRON BLUE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CIBACRON BLUE, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Nuccetelli, M, Mazzetti, A.P, Parker, M.W. | | Deposit date: | 1997-12-16 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
1GWK
 
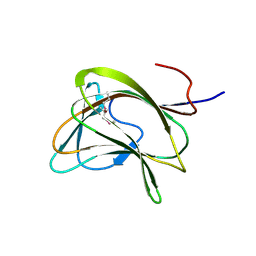 | | Carbohydrate binding module family29 | | Descriptor: | NON-CATALYTIC PROTEIN 1 | | Authors: | Charnock, S.J, Nurizzo, D, Davies, G.J. | | Deposit date: | 2002-03-19 | | Release date: | 2003-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4A9S
 
 | | CRYSTAL STRUCTURE OF HUMAN CHK2 IN COMPLEX WITH BENZIMIDAZOLE CARBOXAMIDE INHIBITOR | | Descriptor: | 2-(4-(3-HYDROXYPHENOXY)PHENYL)-1H-BENZO[D]IMIDAZOLE-5-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Matijssen, C, Silva-Santisteban, M.C, Westwood, I.M, Siddique, S, Choi, V, Sheldrake, P, van Montfort, R.L.M, Blagg, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Benzimidazole Inhibitors of the Protein Kinase Chk2: Clarification of the Binding Mode by Flexible Side Chain Docking and Protein-Ligand Crystallography.
Bioorg.Med.Chem., 20, 2012
|
|
8J19
 
 | | Cryo-EM structure of the LY237-bound GPR84 receptor-Gi complex | | Descriptor: | 6-nonylpyridine-2,4-diol, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J18
 
 | | Cryo-EM structure of the 3-OH-C12-bound GPR84 receptor-Gi complex | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
4RMG
 
 | | Human Sirt2 in complex with SirReal2 and NAD+ | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, NAD-dependent protein deacetylase sirtuin-2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
6CF0
 
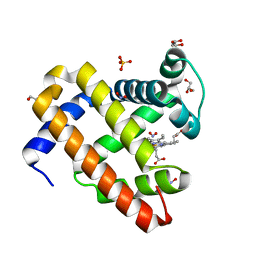 | | Sperm Whale Myoglobin H64V Mutant with Nitrite | | Descriptor: | GLYCEROL, Myoglobin, NITRITE ION, ... | | Authors: | Powell, S.M, Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
1GWL
 
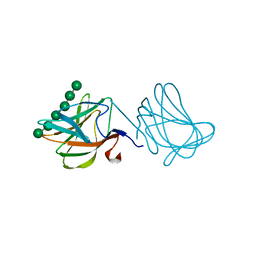 | | Carbohydrate binding module family29 complexed with mannohexaose | | Descriptor: | NON-CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Charnock, S.J, Nurizzo, D, Davies, G.J. | | Deposit date: | 2002-03-19 | | Release date: | 2003-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5KHR
 
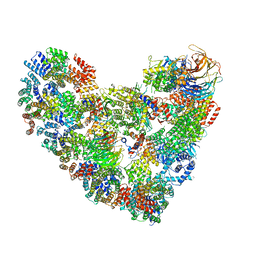 | | Model of human Anaphase-promoting complex/Cyclosome complex (APC15 deletion mutant) in complex with the E2 UBE2C/UBCH10 poised for ubiquitin ligation to substrate (APC/C-CDC20-substrate-UBE2C) | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | VanderLinden, R, Yamaguchi, M, Dube, P, Haselbach, D, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation.
Mol.Cell, 63, 2016
|
|
8B8T
 
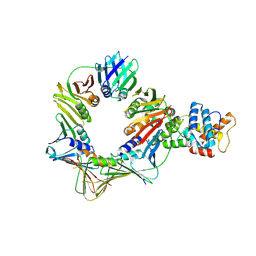 | | Open conformation of the complex of DNA ligase I on PCNA and DNA in the presence of ATP | | Descriptor: | DNA ligase 1, Proliferating cell nuclear antigen | | Authors: | Blair, K, Tehseen, M, Raducanu, V.S, Shahid, T, Lancey, C, Cruehet, R, Hamdan, S, De Biasio, A. | | Deposit date: | 2022-10-05 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of human Lig1 regulation by PCNA in Okazaki fragment sealing.
Nat Commun, 13, 2022
|
|
1AOB
 
 | | E. COLI THYMIDYLATE SYNTHASE COMPLEXED WITH DDURD | | Descriptor: | 2'-5'DIDEOXYURIDINE, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-30 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
1BDU
 
 | |
1GWM
 
 | | Carbohydrate binding module family29 complexed with glucohexaose | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, NON-CATALYTIC PROTEIN 1, ... | | Authors: | Charnock, S.J, Nurizzo, D, Davies, G.J. | | Deposit date: | 2002-03-19 | | Release date: | 2003-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5L97
 
 | |
