2R07
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(6-CHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
2R06
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
1HRI
 
 | | STRUCTURE DETERMINATION OF ANTIVIRAL COMPOUND SCH 38057 COMPLEXED WITH HUMAN RHINOVIRUS 14 | | Descriptor: | 1-[6-(2-CHLORO-4-METHYXYPHENOXY)-HEXYL]-IMIDAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Zhang, A, Nanni, R.G, Oren, D.A, Arnold, E. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of antiviral compound SCH 38057 complexed with human rhinovirus 14.
J.Mol.Biol., 230, 1993
|
|
2KIS
 
 | | Solution structure of CA150 FF1 domain and FF1-FF2 interdomain linker | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Murphy, J.M, Hansen, D, Wiesner, S, Muhandiram, D, Borg, M, Smith, M.J, Sicheri, F, Kay, L.E, Forman-Kay, J.D, Pawson, T. | | Deposit date: | 2009-05-08 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural studies of FF domains of the transcription factor CA150 provide insights into the organization of FF domain tandem arrays.
J.Mol.Biol., 393, 2009
|
|
1RMU
 
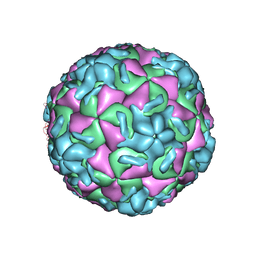 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
2PLV
 
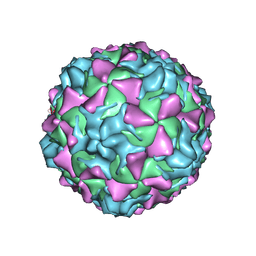 | |
7Z3P
 
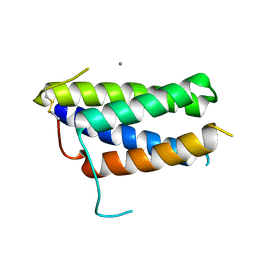 | | Crystal structure of the mouse leptin:LepR-CRH2 encounter complex to 1.95 A resolution. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Leptin, ... | | Authors: | Tsirigotaki, A, Verschueren, K, Savvides, S.N, Verstraete, K. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z3Q
 
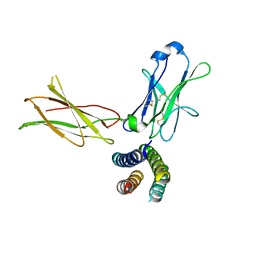 | | Crystal structure of the human leptin:LepR-CRH2 encounter complex to 3.6 A resolution. | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Verschueren, K, Alexandra, T, Savvides, S.N. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.617 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVF
 
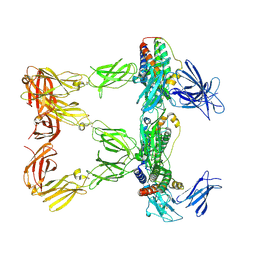 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (closed 3:3 model) | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (6.45 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVO
 
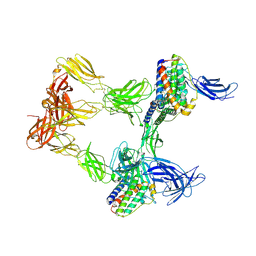 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (open 3:3 model). | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.84 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVE
 
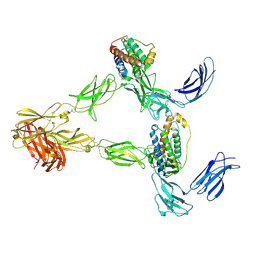 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (2:2 model) | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.62 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8P7G
 
 | |
2EYQ
 
 | |
2I53
 
 | | Crystal structure of Cyclin K | | Descriptor: | ACETATE ION, Cyclin K | | Authors: | Baek, K, Brown, R.S, Birrane, G, Ladias, J.A.A. | | Deposit date: | 2006-08-23 | | Release date: | 2007-01-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human Cyclin K, a Positive Regulator of Cyclin-dependent Kinase 9.
J.Mol.Biol., 366, 2007
|
|
3NS9
 
 | | Crystal structure of CDK2 in complex with inhibitor BS-194 | | Descriptor: | (2S,3S)-3-{[7-(benzylamino)-3-(1-methylethyl)pyrazolo[1,5-a]pyrimidin-5-yl]amino}butane-1,2,4-triol, Cell division protein kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2010-07-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Novel Pyrazolo[1,5-a]pyrimidine Is a Potent Inhibitor of Cyclin-Dependent Protein Kinases 1, 2, and 9, Which Demonstrates Antitumor Effects in Human Tumor Xenografts Following Oral Administration.
J.Med.Chem., 53, 2010
|
|
2A4L
 
 | |
2XPL
 
 | | Crystal structure of Iws1(Spn1) conserved domain from Encephalitozoon cuniculi | | Descriptor: | CHLORIDE ION, IWS1 | | Authors: | Koch, M, Diebold, M.-L, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
3PT2
 
 | | Structure of a viral OTU domain protease bound to Ubiquitin | | Descriptor: | 1.7.6 3-bromanylpropan-1-amine, ACETATE ION, RNA polymerase, ... | | Authors: | James, T.W, Bacik, J.P, Frias-Staheli, N, Garcia-Sastre, A, Mark, B.L. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-19 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7KPF
 
 | | NME2 bound to myristoyl-CoA | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleoside diphosphate kinase B, ... | | Authors: | Price, I.R, Lin, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Chemical Proteomic Approach reveals the regulation of NME1/2 and cancer metastasis by Long-Chain Fatty Acyl Coenzyme A
To Be Published
|
|
1R08
 
 | | STRUCTURAL ANALYSIS OF ANTIVIRAL AGENTS THAT INTERACT WITH THE CAPSID OF HUMAN RHINOVIRUSES | | Descriptor: | 5-(5-(2,6-DICHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PENTYL)-3-(HYDROXYETHYL OXYMETHYLENEOXYMETHYL) ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Smith, T.J, Rossmann, M.G. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of antiviral agents that interact with the capsid of human rhinoviruses.
Proteins, 6, 1989
|
|
3PY1
 
 | | CDK2 ternary complex with SU9516 and ANS | | Descriptor: | (3Z)-3-(1H-IMIDAZOL-5-YLMETHYLENE)-5-METHOXY-1H-INDOL-2(3H)-ONE, 1,2-ETHANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, ... | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
3PXZ
 
 | | CDK2 ternary complex with JWS648 and ANS | | Descriptor: | 2-(4,6-diamino-1,3,5-triazin-2-yl)-4-methoxyphenol, 8-ANILINO-1-NAPHTHALENE SULFONATE, Cell division protein kinase 2 | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
4GAT
 
 | | SOLUTION NMR STRUCTURE OF THE WILD TYPE DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13BP DNA CONTAINING A CGATA SITE, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*GP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors.
J.Mol.Biol., 277, 1998
|
|
6DGP
 
 | |
6DU2
 
 | | Structure of Scp1 D96N bound to REST-pS861/4 peptide | | Descriptor: | MAGNESIUM ION, REST-pS861/4, carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 | | Authors: | Burkholder, N.T, Mayfield, J.E, Yu, X, Irani, S, Arce, D.K, Jiang, F, Matthews, W, Xue, Y, Zhang, Y.J. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST).
J. Biol. Chem., 293, 2018
|
|
