5OFV
 
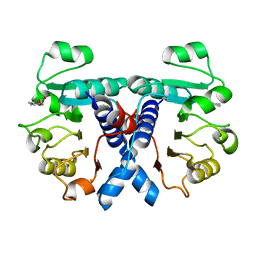 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-fluoro-2-methylbenzoic acid | | Descriptor: | 5-fluoranyl-2-methyl-benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
6CPR
 
 | | Crystal structure of 4-1BBL/4-1BB complex in C2 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Aruna, B, Zajonc, D.M. | | Deposit date: | 2018-03-14 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
5OEW
 
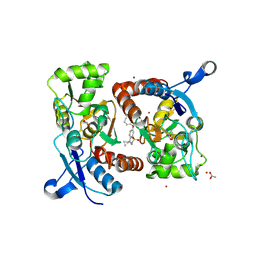 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with glutamate and positive allosteric modulator BPAM538 | | Descriptor: | 4-cyclopropyl-7-(3-methoxyphenoxy)-2,3-dihydro-1$l^{6},2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Larsen, A.P, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-07-10 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 7-Phenoxy-Substituted 3,4-Dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides as Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors with Nanomolar Potency.
J. Med. Chem., 61, 2018
|
|
5NZO
 
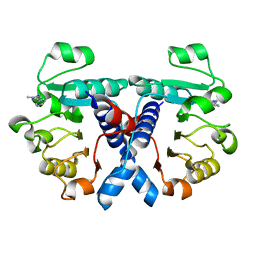 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 1-methyl-3-phenyl-1H-pyrazol-5-amine | | Descriptor: | 2-methyl-5-phenyl-pyrazol-3-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
6BNX
 
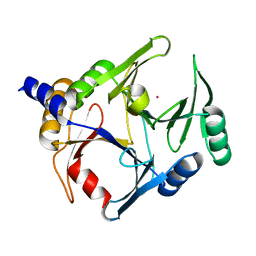 | | Crystal structure of V278E-glyoxalase I mutant from Zea mays in space group P6(3) | | Descriptor: | COBALT (II) ION, Lactoylglutathione lyase | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
5NIH
 
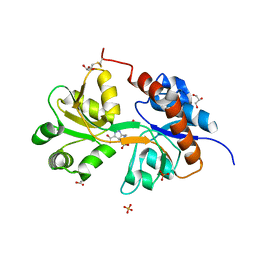 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with agonist LM-12b at 1.3 A resolution. | | Descriptor: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
5NG9
 
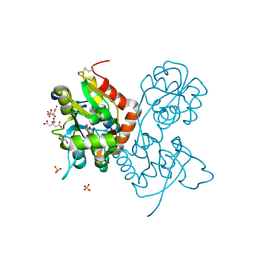 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with agonist CIP-AS at 1.15 A resolution. | | Descriptor: | (2~{S},3~{R},4~{R})-3-(carboxycarbonyl)-4-oxidanyl-pyrrolidine-2-carboxylic acid, (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, CITRATE ANION, ... | | Authors: | Laulumaa, S, Frydenvang, K.A, Winther, S, Kastrup, J.S. | | Deposit date: | 2017-03-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
6CU0
 
 | | Crystal structure of 4-1BBL/4-1BB (C121S) complex in P21 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Aruna, B, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
8ILU
 
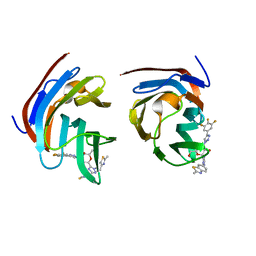 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | (2R,3R,4R,5R,6S)-2-(hydroxymethyl)-6-[2-(2-methyl-1,3-benzothiazol-6-yl)-1,2,4-triazol-3-yl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, SODIUM ION, ... | | Authors: | Kumar, A, Jinal, S, Raman, S, Ghosh, K. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of benzothiazole derived monosaccharides as potent, selective, and orally bioavailable inhibitors of human and mouse galectin-3; a rare example of using a S···O binding interaction for drug design.
Bioorg.Med.Chem., 101, 2024
|
|
5OHQ
 
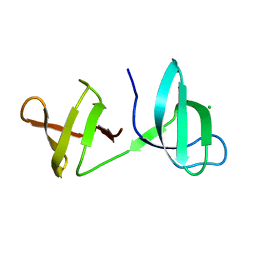 | |
6D3N
 
 | | Crystal structure of h4-1BB ligand | | Descriptor: | GLYCEROL, Tumor necrosis factor ligand superfamily member 9 | | Authors: | Aruna, B, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
5NFF
 
 | | Crystal structure of GP1 receptor binding domain from Morogoro virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Israeli, H, Cohen-Dvashi, H, Shulman, A, Shimon, A, Diskin, R. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other old world arenaviruses.
PLoS Pathog., 13, 2017
|
|
5OFW
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-Chloro-4-fluorobenzamide | | Descriptor: | 3-chloranyl-4-fluoranyl-benzamide, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
6K77
 
 | |
5OHO
 
 | |
5O1T
 
 | |
6CIV
 
 | |
5SYG
 
 | | Cryo-EM reconstruction of zampanolide-bound microtubule | | Descriptor: | (2Z,4E)-N-[(S)-[(1S,2E,5S,8E,10Z,17S)-3,11-dimethyl-19-methylidene-7,13-dioxo-6,21-dioxabicyclo[15.3.1]henicosa-2,8,10-trien-5-yl](hydroxy)methyl]hexa-2,4-dienamide, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kellogg, E.H, Hejab, N.M.A, Nogales, E. | | Deposit date: | 2016-08-11 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Insights into the Distinct Mechanisms of Action of Taxane and Non-Taxane Microtubule Stabilizers from Cryo-EM Structures.
J. Mol. Biol., 429, 2017
|
|
1AD1
 
 | | DIHYDROPTEROATE SYNTHETASE (APO FORM) FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIHYDROPTEROATE SYNTHETASE, POTASSIUM ION | | Authors: | Kostrewa, D, Oefner, C, D'Arcy, A. | | Deposit date: | 1997-02-19 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the dihydropteroate synthase from Staphylococcus aureus.
J.Mol.Biol., 268, 1997
|
|
1AD4
 
 | |
6BNN
 
 | | Crystal structure of V278E-glyoxalase I mutant from Zea mays in space group P4(1)2(1)2 | | Descriptor: | COBALT (II) ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
6TIF
 
 | | ReoM- Listeria monocytogenes | | Descriptor: | SULFATE ION, UPF0297 protein lmo1503 | | Authors: | Rutter, Z.J, Lewis, R.J. | | Deposit date: | 2019-11-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PrkA controls peptidoglycan biosynthesis through the essential phosphorylation of ReoM.
Elife, 9, 2020
|
|
1BCY
 
 | | RECOMBINANT RAT ANNEXIN V, T72K MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BC0
 
 | | RECOMBINANT RAT ANNEXIN V, W185A MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
1BCZ
 
 | | RECOMBINANT RAT ANNEXIN V, T72S MUTANT | | Descriptor: | ANNEXIN V, CALCIUM ION, SULFATE ION | | Authors: | Mo, Y.D, Swairjo, M.A, Li, C.W, Head, J.F, Seaton, B.A. | | Deposit date: | 1998-05-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational and crystallographic analyses of interfacial residues in annexin V suggest direct interactions with phospholipid membrane components.
Biochemistry, 37, 1998
|
|
