5UHY
 
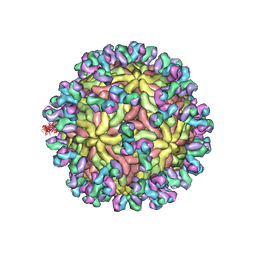 | | A Human Antibody Against Zika Virus Crosslinks the E Protein to Prevent Infection | | Descriptor: | ZV67 Fab chain 1, ZV67 Fab chain 2, envelope protein | | Authors: | Hasan, S.S, Miller, A, Sapparapu, G, Fernandez, E, Klose, T, Long, F, Fokine, A, Porta, J.C, Jiang, W, Diamond, M.S, Crowe Jr, J.E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2017-01-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | A human antibody against Zika virus crosslinks the E protein to prevent infection.
Nat Commun, 8, 2017
|
|
6N5A
 
 | |
1TNT
 
 | |
1TNS
 
 | |
8S6F
 
 | | A Structural Investigation of the Interaction between a GC-376-Based Peptidomimetic PROTAC and Its Precursor with the Viral Main Protease of Coxsackievirus B3 | | Descriptor: | Protease 3C, methyl (4~{S})-4-[[(2~{S})-4-methyl-2-(phenylmethoxycarbonylamino)pentanoyl]amino]-5-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | De Santis, A, Grifagni, D, Orsetti, A, Lenci, E, Rosato, A, D'Onofrio, M, Trabocchi, A, Ciofi-Baffoni, S, Cantini, F, Calderone, V. | | Deposit date: | 2024-02-27 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Structural Investigation of the Interaction between a GC-376-Based Peptidomimetic PROTAC and Its Precursor with the Viral Main Protease of Coxsackievirus B3.
Biomolecules, 14, 2024
|
|
3DS9
 
 | |
6MEM
 
 | |
3DSE
 
 | |
3EIU
 
 | | A second transient position of ATP on its trail to the nucleotide-binding site of subunit B of the motor protein A1Ao ATP synthase | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ADENOSINE-5'-TRIPHOSPHATE, V-type ATP synthase beta chain | | Authors: | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | A second transient position of ATP on its trail to the nucleotide-binding site of subunit B of the motor protein A(1)A(O) ATP synthase
J.Struct.Biol., 166, 2009
|
|
1A8W
 
 | |
3M3J
 
 | | A new crystal form of Lys48-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Ubiquitin | | Authors: | Trempe, J.F, Brown, N.R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new crystal form of Lys48-linked diubiquitin.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2X30
 
 | | Crystal structure of the r139n mutant of a bifunctional enzyme pria | | Descriptor: | PHOSPHORIBOSYL ISOMERASE A, SULFATE ION | | Authors: | Noda-Garcia, L, Camacho-Zarco, A.R, Verdel-Aranda, K, Wright, H, Soberon, X, Fulop, V, Barona-Gomez, F. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and Analysis of Residues Contained on Beta --> Alpha Loops of the Dual-Substrate (Betaalpha)(8) Phosphoribosyl Isomerase a (Pria) Specific for its Phosphoribosyl Anthranilate Isomerase Activity.
Protein Sci., 19, 2010
|
|
5HGG
 
 | | Crystal structure of uPA in complex with a camelid-derived antibody fragment | | Descriptor: | (3S)-3-[(2S,3S,4R)-3,4-DIMETHYLTETRAHYDROFURAN-2-YL]BUTYL LAURATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Camelid Derived Antibody Fragment, ... | | Authors: | Yung, K.W.Y, Kromann-Hansen, T, Andreasen, P.A, Ngo, J.C.K. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Camelid-derived Antibody Fragment Targeting the Active Site of a Serine Protease Balances between Inhibitor and Substrate Behavior
J.Biol.Chem., 291, 2016
|
|
1ADD
 
 | |
1B5Q
 
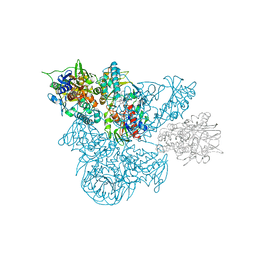 | | A 30 ANGSTROM U-SHAPED CATALYTIC TUNNEL IN THE CRYSTAL STRUCTURE OF POLYAMINE OXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, N,N'-BIS(2,3-BUTADIENYL)-1,4-BUTANE-DIAMINE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase.
Structure Fold.Des., 7, 1999
|
|
6B8U
 
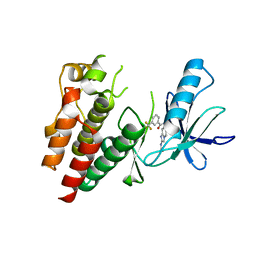 | | Crystals Structure of B-Raf kinase domain in complex with an Imidazopyridinyl benzamide inhibitor | | Descriptor: | Serine/threonine-protein kinase B-raf, ~{N}-[3-(2-acetamidoimidazo[1,2-a]pyridin-6-yl)-4-methyl-phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Appleton, B.A, Murray, J, Shafer, C.M. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Imidazo[1,2-a]pyridin-6-yl-benzamide analogs as potent RAF inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6QMU
 
 | |
3FYI
 
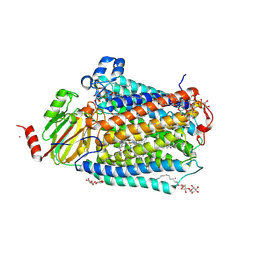 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides in the reduced state bound with cyanide | | Descriptor: | CADMIUM ION, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Qin, L, Mills, D.A, Proshlyakov, D.A, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redox dependent conformational changes in cytochrome c oxidase suggest a gating mechanism for proton uptake.
Biochemistry, 48, 2009
|
|
2MC5
 
 | | A bacteriophage transcription regulator inhibits bacterial transcription initiation by -factor displacement | | Descriptor: | 45L | | Authors: | Liu, B, Shadrin, A, Sheppard, C, Xu, Y, Severinov, K, Matthews, S, Wigneshweraraj, S. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement.
Nucleic Acids Res., 42, 2014
|
|
3NSU
 
 | | A Systematic Screen for Protein-Lipid Interactions in Saccharomyces cerevisiae | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, SULFATE ION | | Authors: | Gallego, O, Fernandez-Tornero, C, Aguilar-Gurrieri, C, Muller, C, Gavin, A.C. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae.
Mol. Syst. Biol., 6, 2010
|
|
3O6B
 
 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) low resolution | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3OAZ
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | CHLORIDE ION, Fab 2G12, heavy chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-06 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OAY
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KQO
 
 | |
4R10
 
 | | A conserved phosphorylation switch controls the interaction between cadherin and beta-catenin in vitro and in vivo | | Descriptor: | 1,2-ETHANEDIOL, Cadherin-related hmr-1, Protein humpback-2, ... | | Authors: | Choi, H.-J, Loveless, T, Lynch, A, Bang, I, Hardin, J, Weis, W.I. | | Deposit date: | 2014-08-03 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Conserved Phosphorylation Switch Controls the Interaction between Cadherin and beta-Catenin In Vitro and In Vivo
Dev.Cell, 33, 2015
|
|
