1YBO
 
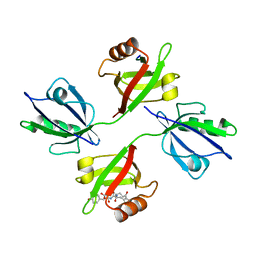 | | Crystal structure of the PDZ tandem of human syntenin with syndecan peptide | | Descriptor: | Syndecan-4, Syntenin 1 | | Authors: | Grembecka, J, Cooper, D.R, Cierpicki, T, Kang, B.S, Devedjiev, Y, Derewenda, Z. | | Deposit date: | 2004-12-21 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The binding of the PDZ tandem of syntenin to target proteins
Biochemistry, 45, 2006
|
|
1Y7N
 
 | | Solution structure of the second PDZ domain of the human neuronal adaptor X11alpha | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 1 | | Authors: | Duquesne, A.E, de Ruijter, M, Brouwer, J, Drijfhout, J.W, Nabuurs, S.B, Spronk, C.A.E.M, Vuister, G.W, Ubbink, M, Canters, G.W. | | Deposit date: | 2004-12-09 | | Release date: | 2005-11-22 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of the neuronal adaptor X11alpha and its interaction with the C-terminal peptide of the human copper chaperone for superoxide dismutase
J.Biomol.Nmr, 32, 2005
|
|
7W6X
 
 | |
4WSI
 
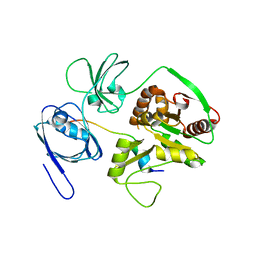 | | Crystal Structure of PALS1/Crb complex | | Descriptor: | MAGUK p55 subfamily member 5, Protein crumbs | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4A9G
 
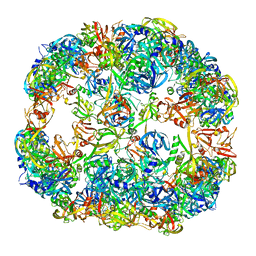 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 24-mer in complex with beta-casein | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8C
 
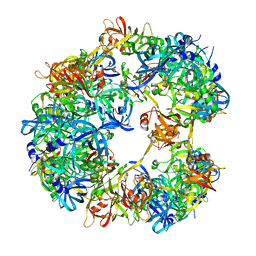 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with a binding peptide | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
5V6T
 
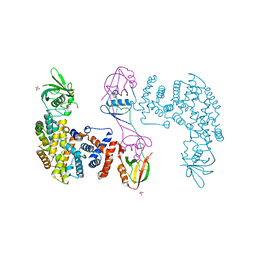 | | The Plexin D1 intracellular region in complex with GIPC1 | | Descriptor: | PDZ domain-containing protein GIPC1, Plexin-D1, SULFATE ION | | Authors: | Shang, G, Zhang, X. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.189 Å) | | Cite: | Structure analyses reveal a regulated oligomerization mechanism of the PlexinD1/GIPC/myosin VI complex.
Elife, 6, 2017
|
|
5WQL
 
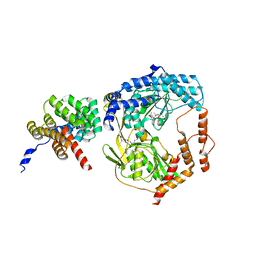 | |
4QL6
 
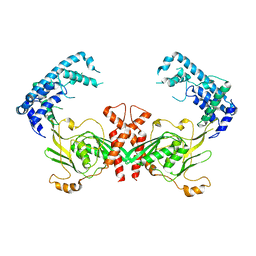 | | Structure of C. trachomatis CT441 | | Descriptor: | Carboxy-terminal processing protease | | Authors: | Kohlmann, F, Hilgenfeld, R, Hansen, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural basis of the proteolytic and chaperone activity of Chlamydia trachomatis CT441
J.Bacteriol., 197, 2015
|
|
3SHW
 
 | | Crystal structure of ZO-1 PDZ3-SH3-Guk supramodule complex with Connexin-45 peptide | | Descriptor: | Gap junction gamma-1 protein, Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
3STJ
 
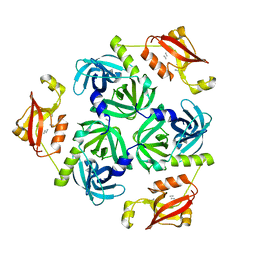 | | Crystal structure of the protease + PDZ1 domain of DegQ from Escherichia coli | | Descriptor: | Protease degQ, peptide (UNK) | | Authors: | Sawa, J, Malet, H, Krojer, T, Canellas, F, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope.
J.Biol.Chem., 286, 2011
|
|
3SUZ
 
 | | Crystal structure of Rat Mint2 PPC | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 2 | | Authors: | Shen, Y, Long, J, Yan, X, Xie, X. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Open-closed motion of Mint2 regulates APP metabolism
J Mol Cell Biol, 5, 2013
|
|
6SAK
 
 | |
3TMH
 
 | | Crystal structure of dual-specific A-kinase anchoring protein 2 in complex with cAMP-dependent protein kinase A type II alpha and PDZK1 | | Descriptor: | A-kinase anchor protein 10, mitochondrial, Na(+)/H(+) exchange regulatory cofactor NHE-RF3, ... | | Authors: | Sarma, G.N, Phan, R.H, Sankaran, B, Taylor, S.S. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Differential mode of D-AKAP2 to PKA isoforms: Insights from D-AKAP2 PKA RII PDZK1 ternary complex structure
To be Published
|
|
3TSZ
 
 | |
3TSW
 
 | | crystal structure of the PDZ3-SH3-GUK core module of Human ZO-1 | | Descriptor: | SULFATE ION, Tight junction protein ZO-1 | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | The Src Homology 3 Domain Is Required for Junctional Adhesion Molecule Binding to the Third PDZ Domain of the Scaffolding Protein ZO-1.
J.Biol.Chem., 286, 2011
|
|
1KY9
 
 | | Crystal Structure of DegP (HtrA) | | Descriptor: | PROTEASE DO | | Authors: | Krojer, T, Garrido-Franco, M, Huber, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2002-02-04 | | Release date: | 2002-04-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine.
Nature, 416, 2002
|
|
6IQQ
 
 | |
6IRD
 
 | | Complex structure of INADL PDZ89 and PLCb4 C-terminal CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, GOLD ION, InaD-like protein | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
6IRE
 
 | | Complex structure of INAD PDZ45 and NORPA CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, Inactivation-no-after-potential D protein | | Authors: | Ye, F, Li, J, Deng, X, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
6IQR
 
 | |
6IQS
 
 | |
6JJL
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-ARG-LYS-LEU, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy.
Commun Biol, 3, 2020
|
|
6JJK
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-TYR-LYS-ILE, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
6JJO
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | Periplasmic serine endoprotease DegP, TMB-CYRKL modulator | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.157 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
