7BCR
 
 | | Crystal structure of the sugar acid binding protein DctPAm from Advenella mimigardefordensis strain DPN7T in complex with galactonate | | Descriptor: | L-galactonic acid, Putative TRAP transporter solute receptor DctP | | Authors: | Schaefer, L, Meinert, C, Kobus, S, Hoeppner, A, Smits, S.H, Steinbuechel, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the sugar acid-binding protein CxaP from a TRAP transporter in Advenella mimigardefordensis strain DPN7 T .
Febs J., 288, 2021
|
|
3V5Z
 
 | | Structure of FBXL5 hemerythrin domain, C2 cell, grown anaerobically | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1847 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
4C7Z
 
 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), activated with sodium dithionite and sodium sulfide | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Kinetic and Structural Studies of Aldehyde Oxidoreductase from Desulfovibrio Gigas Reveal a Dithiolene-Based Chemistry for Enzyme Activation and Inhibition by H2O2.
Plos One, 8, 2013
|
|
4C80
 
 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with hydrogen peroxide | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic and Structural Studies of Aldehyde Oxidoreductase from Desulfovibrio Gigas Reveal a Dithiolene-Based Chemistry for Enzyme Activation and Inhibition by H2O2.
Plos One, 8, 2013
|
|
4C7Y
 
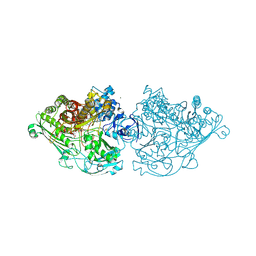 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with sodium dithionite and sodium sulfide | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Kinetic and Structural Studies of Aldehyde Oxidoreductase from Desulfovibrio Gigas Reveal a Dithiolene-Based Chemistry for Enzyme Activation and Inhibition by H2O2.
Plos One, 8, 2013
|
|
6R7N
 
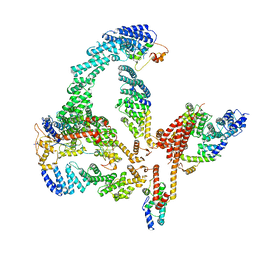 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7H
 
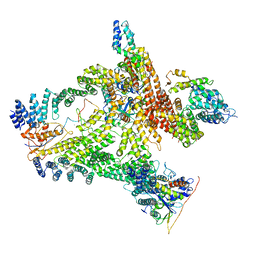 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6VR8
 
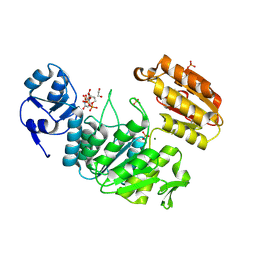 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mur ligase middle domain protein, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2020-02-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
1ZAU
 
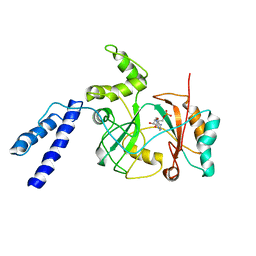 | |
2MXS
 
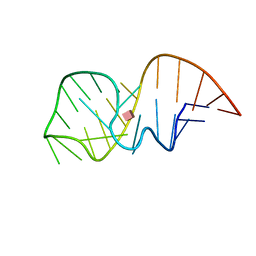 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | Descriptor: | PAROMOMYCIN, RNA (27-MER) | | Authors: | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | Deposit date: | 2015-01-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5XMJ
 
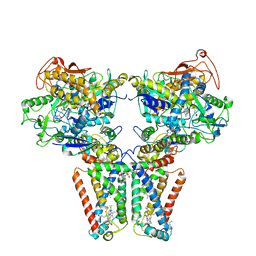 | | Crystal structure of quinol:fumarate reductase from Desulfovibrio gigas | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Guan, H.H, Hsieh, Y.C, Lin, P.R, Chen, C.J. | | Deposit date: | 2017-05-15 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas.
Sci Rep, 8, 2018
|
|
6TTU
 
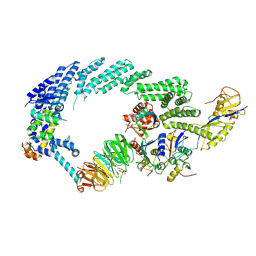 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | Descriptor: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
2OWO
 
 | | Last Stop on the Road to Repair: Structure of E.coli DNA Ligase Bound to Nicked DNA-Adenylate | | Descriptor: | 26-MER, 5'-D(*AP*CP*AP*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*C)-3', 5'-D(*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*G)-3', ... | | Authors: | Shuman, S, Nandakumar, J, Nair, P.A. | | Deposit date: | 2007-02-16 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Last Stop on the Road to Repair: Structure of E. coli DNA Ligase Bound to Nicked DNA-Adenylate.
Mol.Cell, 26, 2007
|
|
6NHX
 
 | | mycobacterial DNA ligase D complexed with ATP and MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent DNA ligase | | Authors: | Shuman, S, Unciuleac, M, Goldgur, Y. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of ATP-bound DNA ligase D in a closed domain conformation reveal a network of amino acid and metal contacts to the ATP phosphates.
J. Biol. Chem., 294, 2019
|
|
2CFM
 
 | |
8IVI
 
 | | crystal structure of a medium-long chain fatty acyl-CoA ligase | | Descriptor: | Medium/long-chain-fatty-acid--CoA ligase FadD8 | | Authors: | Li, S. | | Deposit date: | 2023-03-27 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural study of medium-long chain fatty acyl-CoA ligase FadD8 from Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
6NHZ
 
 | | mycobacterial DNA ligase D complexed with ATP and Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent DNA ligase, MAGNESIUM ION | | Authors: | Shuman, S, Unciuleac, M, Goldgur, Y. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of ATP-bound DNA ligase D in a closed domain conformation reveal a network of amino acid and metal contacts to the ATP phosphates.
J. Biol. Chem., 294, 2019
|
|
5XMC
 
 | |
2DTI
 
 | |
5D1P
 
 | | Archaeal ATP-dependent RNA ligase - form 2 | | Descriptor: | ATP-dependent RNA ligase, MAGNESIUM ION, SULFATE ION | | Authors: | Murakami, K.S. | | Deposit date: | 2015-08-04 | | Release date: | 2016-03-09 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis.
Nucleic Acids Res., 44, 2016
|
|
2DKG
 
 | |
6MKB
 
 | | Crystal structure of murine 4-1BB ligand | | Descriptor: | SODIUM ION, SULFATE ION, Tumor necrosis factor ligand superfamily member 9, ... | | Authors: | Bitra, A, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the m4-1BB/4-1BBL complex reveals an unusual dimeric ligand that undergoes structural changes upon 4-1BB receptor binding.
J. Biol. Chem., 294, 2019
|
|
6DT1
 
 | | Crystal structure of the ligase from bacteriophage T4 complexed with DNA intermediate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction.
Nucleic Acids Res., 46, 2018
|
|
7KR4
 
 | |
3L2P
 
 | | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching Between Two DNA Bound States | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*CP*CP*G)-3', 5'-D(*GP*TP*CP*GP*GP*AP*CP*TP*G)-3', 5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*C)-3', ... | | Authors: | Cotner-Gohara, E.A, Kim, I.K, Hammel, M, Tainer, J.A, Tomkinson, A, Ellenberger, T. | | Deposit date: | 2009-12-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching between Two DNA-Bound States.
Biochemistry, 49, 2010
|
|
