5A19
 
 | | The structure of MAT2A in complex with PPNP. | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murray, B, Antonyuk, S.V, Marina, A, Lu, S.C, Mato, J.M, Hasnain, S.S, Rojas, A.L. | | Deposit date: | 2015-04-28 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystallography Captures Catalytic Steps in Human Methionine Adenosyltransferase Enzymes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6SZG
 
 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GR839 and GSK513 | | Descriptor: | (4-chlorophenyl)-[(3~{S})-3-oxidanylpiperidin-1-yl]methanone, 4,5,6,7-tetrahydro-2~{H}-indazole-3-carboxylic acid, CALCIUM ION, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6K9H
 
 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{S},3~{S})-2-oxidanylidene-2'-phenyl-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
5ZBK
 
 | |
9FSW
 
 | | ClpP from Staphylococcus epidermidis with glycerol in some of the catalytic sites. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, GLYCEROL | | Authors: | Alves Franca, B, Rohde, H, Betzel, C. | | Deposit date: | 2024-06-22 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular insights into the dynamic modulation of bacterial ClpP function and oligomerization by peptidomimetic boronate compounds.
Sci Rep, 14, 2024
|
|
5EP7
 
 | |
6JX1
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101 | | Descriptor: | Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
4XJ3
 
 | | Crystal structure of Vibrio cholerae DncV GTP bound form | | Descriptor: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
5ALM
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | (3R)-1-(cyclopropylmethyl)-3-methyl-3-(4-methylsulfanylphenoxy)pyrrolidine, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
4XHB
 
 | |
4XJL
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with a HTS lead compound | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, DI(HYDROXYETHYL)ETHER, N-(1,2,3-benzothiadiazol-5-yl)-4-phenylpiperazine-1-carboxamide, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2015-01-08 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment based inhibitor design of Mycobacterium tuberculosis BioA (http://hdl.handle.net/11299/171084)
Thesis, University Of Minnesota, 2015
|
|
3HHM
 
 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha and the drug wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5A6O
 
 | | Crystal structure of the apo form of the unphosphorylated human death associated protein kinase 3 (DAPK3) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AKL
 
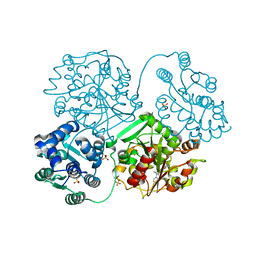 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | BIFUNCTIONAL EPOXIDE HYDROLASE 2, N-(3,3-DIPHENYLPROPYL)PYRROLIDINE-1-CARBOXAMIDE, SULFATE ION | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5RCC
 
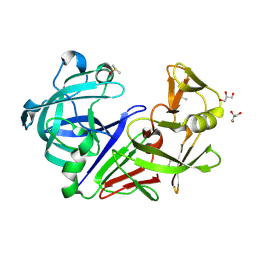 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library H02b | | Descriptor: | 3-cyclopentyl-1-(piperazin-1-yl)propan-1-one, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6K10
 
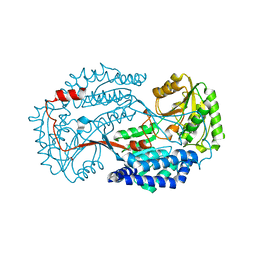 | | Non substrate bound state of Staphylococcus Aureus AldH | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase | | Authors: | Zhang, Z, Tao, X. | | Deposit date: | 2019-05-08 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78962183 Å) | | Cite: | Structural Insight into the Substrate Gating Mechanism by Staphylococcus aureus Aldehyde Dehydrogenase
CCS Chemistry, 2, 2020
|
|
9I08
 
 | | The Paulinella chromatophore transit peptide part2 (crTPpart2) from ArgC | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Klimenko, V, Reiners, J, Applegate, V, Reimann, K, Hoeppner, A, Smits, S.H.J, Nowack, E.C.M. | | Deposit date: | 2025-01-14 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Paulinella chromatophore transit peptide part2 adopts a structural fold similar to the gamma-glutamyl-cyclotransferase fold.
Plant Physiol., 199, 2025
|
|
4BKY
 
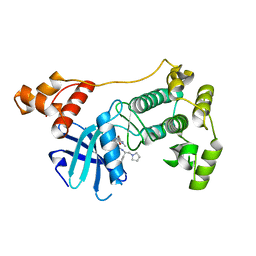 | | Crystal structure of unphosphorylated Maternal Embryonic Leucine zipper Kinase (MELK) in complex with pyrrolopyrazole inhibitor | | Descriptor: | 3'-{[(4-bromo-1-methyl-1H-pyrrol-2-yl)carbonyl]amino}-N-[(1S)-1-phenyl-2-(pyrrolidin-1-yl)ethyl]-1',4'-dihydro-5'H-spiro[cyclopropane-1,6'-pyrrolo[3,4-c]pyrazole]-5'-carboxamide, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE, UNKNOWN ATOM OR ION | | Authors: | Canevari, G, Re Depaolini, S, Cucchi, U, Forte, B, Carpinelli, P, Bertrand, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insight Into Maternal Embryonic Leucine Zipper Kinase (Melk) Conformation and Inhibition Towards Structure- Based Drug Design.
Biochemistry, 52, 2013
|
|
6DJK
 
 | |
3EF2
 
 | | Structure of the Marasmius oreades mushroom lectin (MOA) in complex with Galalpha(1,3)[Fucalpha(1,2)]Gal and Calcium. | | Descriptor: | ACETATE ION, Agglutinin, CALCIUM ION, ... | | Authors: | Grahn, E.M, Goldstein, I.J, Krengel, U. | | Deposit date: | 2008-09-08 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization of a Lectin from the Mushroom Marasmius oreades in Complex with the Blood Group B Trisaccharide and Calcium.
J.Mol.Biol., 390, 2009
|
|
7TB9
 
 | | Structural characterization of the biological synthetic peptide pCEMP1 | | Descriptor: | CEMP1-p1 | | Authors: | Lopez Giraldo, A, del Rio Portilla, F, Nidome Campos, M, Romo Arevalo, E, Arzate, H. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of cementum protein 1 derived peptide (CEMP1-p1) and its role in the mineralization process.
J.Pept.Sci., 29, 2023
|
|
9QUD
 
 | | Cu(II)-bound de novo protein scaffold TFD-EH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Wagner Egea, P, Delhommel, F, Mustafa, G, Leiss-Maier, F, Klimper, L, Badmann, T, Heider, A, Wille, I.C, Groll, M, Sattler, M, Zeymer, C. | | Deposit date: | 2025-04-10 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modular protein scaffold architecture and AI-guided sequence optimization facilitate de novo metalloenzyme engineering.
Structure, 2025
|
|
5WZS
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 8 | | Descriptor: | 2-[2-methyl-1-(naphthalen-1-ylmethyl)-3-oxamoyl-indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5RZA
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z729352906 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-fluorophenyl)methyl]-1lambda~6~,2-thiazolidine-1,1-dione, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
6DQO
 
 | |
