3G23
 
 | |
7H66
 
 | |
2X3L
 
 | | Crystal Structure of the Orn_Lys_Arg decarboxylase family protein SAR0482 from Methicillin-resistant Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, ORN/LYS/ARG DECARBOXYLASE FAMILY PROTEIN, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
7H63
 
 | | THE 1.65 A CRYSTAL STRUCTURE OF HUMAN CHYMASE IN COMPLEX WITH 4-[(5-fluoro-3-propan-2-yl-1H-indol-2-yl)-phenylmethyl]-3-hydroxy-2-propan-2-yl-1,2-dihydropyrrol-5-one (VINYLOGOUS ACID) | | Descriptor: | (5S)-3-[(S)-[5-fluoro-3-(propan-2-yl)-1H-indol-2-yl](phenyl)methyl]-4-hydroxy-5-(propan-2-yl)-1,5-dihydro-2H-pyrrol-2-one, Chymase, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D.W, Benz, J.M, Schlatter, D, Hilpert, H. | | Deposit date: | 2024-04-19 | | Release date: | 2025-03-05 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of human Chymase and Cathepsin G
To be published
|
|
6DVU
 
 | |
7H62
 
 | |
6FPN
 
 | | Lytic transglycosylase in action | | Descriptor: | Putative soluble lytic murein transglycosylase, SULFATE ION | | Authors: | Williams, A.H, Hoauz, A, Boneca, I.G. | | Deposit date: | 2018-02-11 | | Release date: | 2018-03-14 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A step-by-stepin crystalloguide to bond cleavage and 1,6-anhydro-sugar product synthesis by a peptidoglycan-degrading lytic transglycosylase.
J. Biol. Chem., 293, 2018
|
|
6IL3
 
 | | Crystal structure of the FLT3 kinase bound to a small molecule inhibitor | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, 7-methoxy-6-(1-methyl-1H-pyrazol-4-yl)-3-(pyridin-2-yl)imidazo[1,2-a]pyridine, CHLORIDE ION, ... | | Authors: | Thomas, C.J. | | Deposit date: | 2018-10-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the FLT3 kinase bound to a small molecule inhibitor
To Be Published
|
|
3GE7
 
 | | tRNA-guanine transglycosylase in complex with 6-amino-4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2009-02-25 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase
Chemmedchem, 4, 2009
|
|
1AP1
 
 | |
6S98
 
 | |
6VD5
 
 | | Crystal Structure of Dehaloperoxidase B in Complex with cofactor Iron(III) Mesoporphyrin IX and Substrate Trichlorophenol | | Descriptor: | 1,2-ETHANEDIOL, 2,4,6-trichlorophenol, Dehaloperoxidase B, ... | | Authors: | Ghiladi, R.A, de Serrano, V.S, Malewschik, T. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Nonnative Heme Incorporation into Multifunctional Globin Increases Peroxygenase Activity an Order and Magnitude Compared to Native Enzyme
To Be Published
|
|
1K09
 
 | | Solution structure of BetaCore, A Designed Water Soluble Four-Stranded Antiparallel b-sheet Protein | | Descriptor: | (7E)-4,9-dioxo-6-oxa-3,7,10-triazadodec-7-ene-1,12-dioic acid, Core Module I, Core Module II | | Authors: | Carulla, N, Woodward, C, Barany, G. | | Deposit date: | 2001-09-18 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-21 | | Method: | SOLUTION NMR | | Cite: | BetaCore, a designed water soluble four-stranded antiparallel beta-sheet protein.
Protein Sci., 11, 2002
|
|
3JSG
 
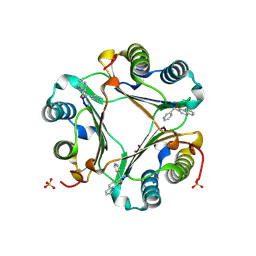 | |
6C5B
 
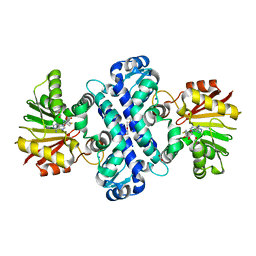 | | Crystal Structure Analysis of LaPhzM | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, FORMIC ACID, Methyltransferase, ... | | Authors: | Beltran, D.G, Schacht, A, Zhang, L. | | Deposit date: | 2018-01-15 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Functional and Structural Analysis of Phenazine O-Methyltransferase LaPhzM from Lysobacter antibioticus OH13 and One-Pot Enzymatic Synthesis of the Antibiotic Myxin.
ACS Chem. Biol., 13, 2018
|
|
3FUP
 
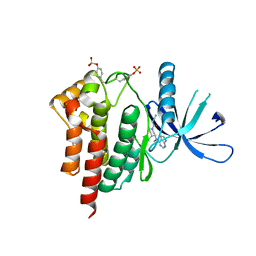 | | Crystal structures of JAK1 and JAK2 inhibitor complexes | | Descriptor: | 3-{(3R,4R)-4-methyl-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}-3-oxopropanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Williams, N.K, Bamert, R.S, Patel, O, Fantino, E, Rossjohn, J, Lucet, I.S. | | Deposit date: | 2009-01-14 | | Release date: | 2009-02-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains.
J.Mol.Biol., 387, 2009
|
|
6S9W
 
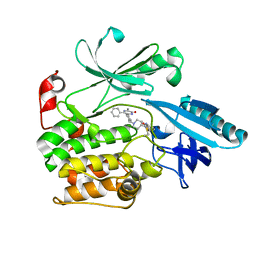 | |
6SAN
 
 | | SALSA / DMBT1 / GP340 SRCR domain 8 soaked in calcium and magnesium | | Descriptor: | CHLORIDE ION, Deleted in malignant brain tumors 1 protein, GLYCEROL, ... | | Authors: | Reichhardt, M.P, Johnson, S, Loimaranta, V, Lea, S.M. | | Deposit date: | 2019-07-17 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold.
Life Sci Alliance, 3, 2020
|
|
6UWY
 
 | | DYRK1A bound to a harmine derivative | | Descriptor: | 4-(7-methoxy-1-methyl-9H-beta-carbolin-9-yl)butanamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, TETRAETHYLENE GLYCOL | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2019-11-05 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis and Biological Validation of a Harmine-Based, Central Nervous System (CNS)-Avoidant, Selective, Human beta-Cell Regenerative Dual-Specificity Tyrosine Phosphorylation-Regulated Kinase A (DYRK1A) Inhibitor.
J.Med.Chem., 63, 2020
|
|
5FPS
 
 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 3-aminobenzene-1,2-dicarboxylic acid (AT1246) in an alternate binding site. | | Descriptor: | 3-AMINOBENZENE-1,2-DICARBOXYLIC ACID, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3JY6
 
 | | Crystal structure of LacI Transcriptional regulator from Lactobacillus brevis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Transcriptional regulator, ... | | Authors: | Syed Ibrahim, B, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of LacI Transcriptional regulator from Lactobacillus brevis
To be Published
|
|
4NRD
 
 | |
7YDQ
 
 | | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | | Descriptor: | 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide, Nucleoside transporter 1,Green fluorescent protein | | Authors: | Wang, C, Yu, L.Y, Li, J.L, Ren, R.B, Deng, D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
2XH6
 
 | | Clostridium perfringens enterotoxin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, HEAT-LABILE ENTEROTOXIN B CHAIN, octyl beta-D-glucopyranoside | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, MCClane, B.A, Basak, A.K. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
6C7R
 
 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
