8GK9
 
 | |
8GBA
 
 | |
8GBM
 
 | |
8GB9
 
 | |
8GBO
 
 | |
8GIV
 
 | |
8FUV
 
 | |
8FVG
 
 | |
8FRS
 
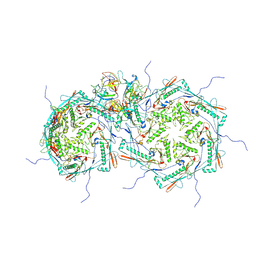 | |
8FVH
 
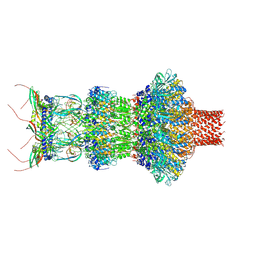 | | Pseudomonas phage E217 neck (portal, head-to-tail connector, collar and gateway proteins) | | Descriptor: | E217 collar protein gp28, E217 gateway protein gp29, E217 head-to-tail connector protein gp27, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2023-01-18 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
8UBZ
 
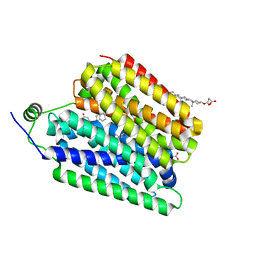 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UBX
 
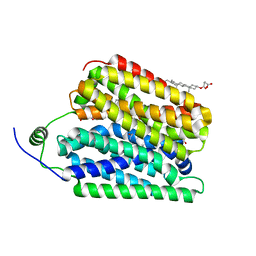 | | Ethanolamine-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, ETHANOLAMINE, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UC0
 
 | | Endogenous ligand bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UBW
 
 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
7MKE
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 2) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKI
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR (-5G to C) promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7YCA
 
 | | Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
7M2K
 
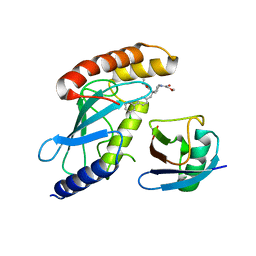 | | CDC34A-Ubiquitin-2ab inhibitor complex | | Descriptor: | 4-[(3',5'-dichloro[1,1'-biphenyl]-4-yl)methyl]-N-ethyl-1-(methoxyacetyl)piperidine-4-carboxamide, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, St-Cyr, D, Tyers, M, Sicheri, F. | | Deposit date: | 2021-03-16 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification and optimization of molecular glue compounds that inhibit a noncovalent E2 enzyme-ubiquitin complex.
Sci Adv, 7, 2021
|
|
8UBY
 
 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
7TX7
 
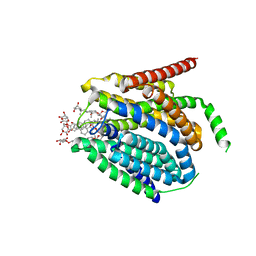 | |
7TX6
 
 | | Cryo-EM structure of the human reduced folate carrier in complex with methotrexate | | Descriptor: | Digitonin, METHOTREXATE, Reduced folate transporter,Soluble cytochrome b562 | | Authors: | Wright, N.J, Fedor, J.G, Lee, S.-Y. | | Deposit date: | 2022-02-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Methotrexate recognition by the human reduced folate carrier SLC19A1.
Nature, 609, 2022
|
|
7U0M
 
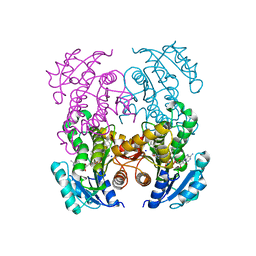 | |
8HIJ
 
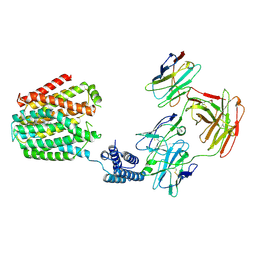 | | The 5-MTHF-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
7URG
 
 | |
8UW1
 
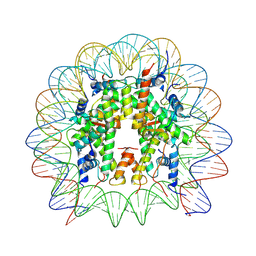 | | Cryo-EM structure of DNMT3A1 UDR in complex with H2AK119Ub-nucleosome | | Descriptor: | DNA (146-MER), DNA (cytosine-5)-methyltransferase 3A, Histone H2A, ... | | Authors: | Gretarsson, K, Abini-Agbomson, S, Armache, K.-J, Lu, C. | | Deposit date: | 2023-11-05 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch.
Sci Adv, 10, 2024
|
|
