8F9W
 
 | | Crystal structure of Ky15.8 Fab in complex with circumsporozoite protein NPDP peptide | | Descriptor: | 1,2-ETHANEDIOL, Circumsporozoite protein NPDP peptide, Ky15.8 Antibody, ... | | Authors: | Thai, E, Prieto, K, Julien, J.P. | | Deposit date: | 2022-11-24 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular determinants of cross-reactivity and potency by VH3-33 antibodies against the Plasmodium falciparum circumsporozoite protein.
Cell Rep, 42, 2023
|
|
9F2X
 
 | | Crystal structure of SARS-CoV-2 Mpro in complex with RHTCR03 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[4-[(2~{S})-1-[[(2~{S},3~{R})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-3-oxidanylidene-pyrazin-2-yl]carbamate | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-24 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
6AXE
 
 | |
3O12
 
 | | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein YJL217W | | Authors: | Zhang, R, Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a functionally unknown protein from Saccharomyces cerevisiae.
TO BE PUBLISHED
|
|
4TU7
 
 | | Structure of U2AF65 D231V variant with BrU5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*UP*U)-3'), GLYCEROL, ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2014-06-24 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5FLY
 
 | | The FhuD protein from S.pseudintermedius | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Malito, E. | | Deposit date: | 2015-10-29 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal Structure of Fhud at 1.6 Angstrom Resolution: A Ferrichrome-Binding Protein from the Animal and Human Pathogen Staphylococcus Pseudintermedius
Acta Crystallogr.,Sect.F, 72, 2016
|
|
9FJX
 
 | |
6IIK
 
 | | USP14 catalytic domain with IU1 | | Descriptor: | 1-[1-(4-fluorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(pyrrolidin-1-yl)ethan-1-one, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Mei, Z.Q, Wang, Y.W, He, W, Wang, F. | | Deposit date: | 2018-10-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade.
Cell Res., 28, 2018
|
|
5LAW
 
 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 14 | | Descriptor: | 2-[(3~{S},3'~{a}~{S},6'~{S},6'~{a}~{S})-6-chloranyl-6'-(3-chlorophenyl)-4'-(cyclopropylmethyl)-2-oxidanylidene-spiro[1~{H}-indole-3,5'-3,3~{a},6,6~{a}-tetrahydro-2~{H}-pyrrolo[3,2-b]pyrrole]-1'-yl]ethanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5FEN
 
 | |
6QAL
 
 | | ERK2 mini-fragment binding | | Descriptor: | 1,1-bis(oxidanylidene)thietan-3-ol, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
7UL4
 
 | | CryoEM Structure of Inactive MOR Bound to Alvimopan and Mb6 | | Descriptor: | Megabody 6, Mu-type opioid receptor, N-[(2S)-2-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]methyl}-3-phenylpropanoyl]glycine | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UOA
 
 | | MAGEA4-MTP1 linear peptide complex | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MTP-1, ... | | Authors: | Williams, R.S, Tumbale, P.S. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery and Structural Basis of the Selectivity of Potent Cyclic Peptide Inhibitors of MAGE-A4.
J.Med.Chem., 65, 2022
|
|
6PVZ
 
 | |
9FK0
 
 | | LGTV with TBEV prME | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Bisikalo, K, Rosendal, E. | | Deposit date: | 2024-06-01 | | Release date: | 2024-11-06 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Influence of the pre-membrane and envelope proteins on structure, pathogenicity, and tropism of tick-borne encephalitis virus.
J.Virol., 99, 2025
|
|
7IEW
 
 | | Crystal structure of Endothiapepsin in complex with fragment EOS102818 from ECBL-96 | | Descriptor: | 1-(quinolin-3-yl)methanamine, Endothiapepsin | | Authors: | Wollenhaupt, J, Benz, L.S, Jirgensons, A, Miletic, T, Mueller, U, Weiss, M.S. | | Deposit date: | 2025-06-05 | | Release date: | 2025-11-05 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | From fragments to follow-ups: rapid hit expansion by making use of EU-OPENSCREEN resources.
Rsc Med Chem, 2025
|
|
4Y9H
 
 | | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, DECANE, DODECANE, ... | | Authors: | Saiki, H, Sugiyama, S, Kakinouchi, K, Kawatake, S, Hanashima, S, Matsumori, N, Murata, M. | | Deposit date: | 2015-02-17 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The 1.43 angstrom crystal structure of bacteriorhodopsin crystallized from bicelles
To Be Published
|
|
4TJV
 
 | | Crystal structure of protease-associated domain of Arabidopsis vacuolar sorting receptor 1 | | Descriptor: | IODIDE ION, Vacuolar-sorting receptor 1 | | Authors: | Luo, F, Fong, Y.H, Jiang, L.W, Wong, K.B. | | Deposit date: | 2014-05-25 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | How vacuolar sorting receptor proteins interact with their cargo proteins: crystal structures of apo and cargo-bound forms of the protease-associated domain from an Arabidopsis vacuolar sorting receptor.
Plant Cell, 26, 2014
|
|
5AMT
 
 | | Intracellular growth locus protein E | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, IGLE | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Cloning, Expression, Purification, Crystallization and Preliminary X-Ray Diffraction Analysis of Intracellular Growth Locus E (Igle) Protein from Francisella Tularensis Subsp. Novicida.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6M4T
 
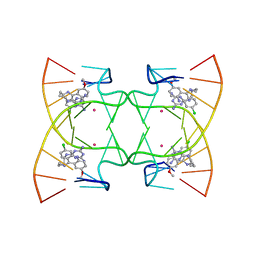 | | U shaped head to head four-way junction in d(TTCTGCTGCTGAA) sequence | | Descriptor: | COBALT (II) ION, DNA (5'-D(P*(UD)P*TP*CP*TP*GP*CP*TP*GP*CP*TP*GP*AP*A)-3'), N4-[4-[(6-chloranyl-2-methoxy-acridin-9-yl)amino]butyl]-1,3,5-triazine-2,4,6-triamine | | Authors: | Hou, M.H, Chien, C.M, Satange, R.B, Wu, P.C. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Targeting T:T Mismatch with Triaminotriazine-Acridine Conjugate Induces a U-Shaped Head-to-Head Four-Way Junction in CTG Repeat DNA.
J.Am.Chem.Soc., 142, 2020
|
|
7KIE
 
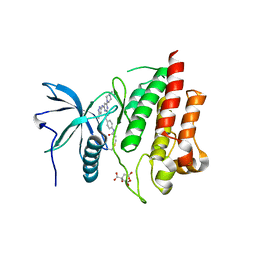 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 3 | | Descriptor: | CITRATE ANION, Fibroblast growth factor receptor 2, GLYCEROL, ... | | Authors: | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
7KL0
 
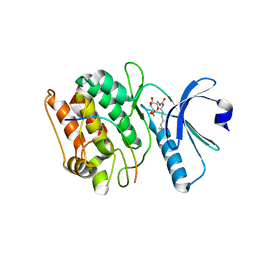 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B(S1303D) | | Descriptor: | 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase type II subunit alpha, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2020-10-28 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
9JUY
 
 | | X-ray crystal structure of Y16513 in CBP | | Descriptor: | (5~{S})-1-(3-chloranyl-4-methoxy-phenyl)-5-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{S})-2-morpholin-4-ylpropyl]imidazol-2-yl]pyrrolidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE | | Authors: | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | Deposit date: | 2024-10-08 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
6I1S
 
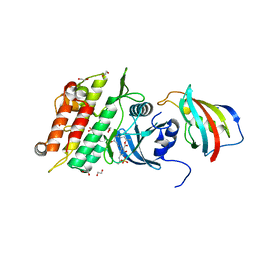 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with FKBP12 and the inhibitor E6201 | | Descriptor: | (4~{S},5~{R},6~{Z},9~{S},10~{S},12~{E})-16-(ethylamino)-4,5-dimethyl-9,10,18-tris(oxidanyl)-3-oxabicyclo[12.4.0]octadeca-1(14),6,12,15,17-pentaene-2,8-dione, 1,2-ETHANEDIOL, Activin receptor type-1, ... | | Authors: | Williams, E.P, Pinkas, D.M, Fortin, J, Newman, J.A, Bradshaw, W.J, Mahajan, P, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-10-30 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas.
Cancer Cell, 37, 2020
|
|
8YD9
 
 | | Crystal structure of p38alpha with an allosteric inhibitor 3 | | Descriptor: | 13-[4-({Imidazo[1,2-a]pyridin-2-yl}methoxy)phenyl]-4,8-dioxa-12,14,16,18-tetraazatetracyclo[9.7.0.0^{3,9}.0^{12,17}]octadeca-1(11),2,9,15,17-pentaen-15-amine, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Hasegawa, S, Kinoshita, T. | | Deposit date: | 2024-02-19 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Identification of a novel target site for ATP-independent p38 alpha inhibitors
To Be Published
|
|
