7EDK
 
 | |
8A7Z
 
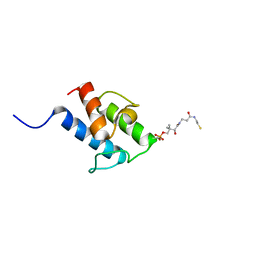 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid polyketide synthase-non ribosomal peptide synthetase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
9ATN
 
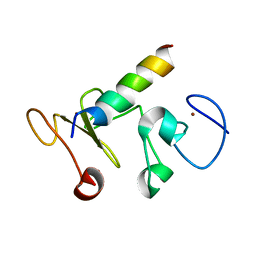 | |
8IJC
 
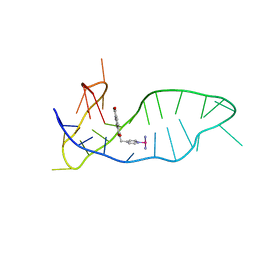 | | NMR solution structure of the 1:1 complex of a platinum(II) ligand L1-transpt covalently bound to a G-quadruplex MYT1L | | Descriptor: | G-quadruplex DNA MYT1L, Pt(NH3)2(2-(pyridin-4-ylmethyl)benzo-[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Mao, Z.-W. | | Deposit date: | 2023-02-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Organic-Platinum Hybrids for Covalent Binding of G-Quadruplexes: Structural Basis and Application to Cancer Immunotherapy.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8K6Z
 
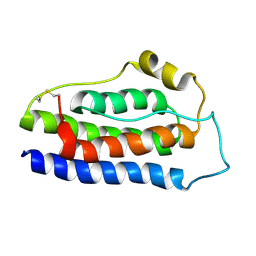 | | NMR structure of human leptin | | Descriptor: | Leptin | | Authors: | Fan, X, Qin, R, Yuan, W, Fan, J, Huang, W, Lin, Z. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Structure, 32, 2024
|
|
6WUX
 
 | |
1Q8G
 
 | | NMR structure of human Cofilin | | Descriptor: | Cofilin, non-muscle isoform | | Authors: | Pope, B.J, Zierler-Gould, K.M, Kuhne, R, Weeds, A.G, Ball, L.J. | | Deposit date: | 2003-08-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human cofilin: rationalizing actin binding and pH sensitivity
J.Biol.Chem., 279, 2004
|
|
7TUJ
 
 | | NMR solution structure of the phosphorylated MUS81-binding region from human SLX4 | | Descriptor: | Structure-specific endonuclease subunit SLX4 | | Authors: | Payliss, B.J, Reichheld, S.E, Lemak, A, Arrowsmith, C.H, Sharpe, S, Wyatt, H.D.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-09 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease.
Cell Rep, 41, 2022
|
|
8HPB
 
 | |
8HQB
 
 | | NMR Structure of OsCIE1-Ubox | | Descriptor: | U-box domain-containing protein 12 | | Authors: | Zhang, Y, Yu, C.Z, Lan, W.X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
7X3A
 
 | | NMR solution structure of the 1:1 complex of a pyridostatin (PDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-azanylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
7X2Z
 
 | | NMR solution structure of the 1:1 complex of a pyridostatin derivative (PyPDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-pyrrolidin-1-ylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-26 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
6MNL
 
 | |
8RAJ
 
 | | NMR structure of PKS docking domains | | Descriptor: | Beta-ketoacyl synthase, Trimethylamine monooxygenase | | Authors: | Scat, S, Weissman, K.J, Chagot, B. | | Deposit date: | 2023-12-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | SOLUTION NMR | | Cite: | Insights into docking in megasynthases from the investigation of the toblerol trans -AT polyketide synthase: many alpha-helical means to an end.
Rsc Chem Biol, 5, 2024
|
|
6VNZ
 
 | |
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
5IIR
 
 | |
5IEW
 
 | |
7OFN
 
 | |
7XGA
 
 | |
7XFG
 
 | |
7XEZ
 
 | |
6OBK
 
 | |
6OWR
 
 | | NMR solution structure of YfiD | | Descriptor: | Autonomous glycyl radical cofactor | | Authors: | Bowman, S.E.J, Drennan, C.L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biochemical characterization of a spare part protein that restores activity to an oxygen-damaged glycyl radical enzyme.
J.Biol.Inorg.Chem., 24, 2019
|
|
5VSO
 
 | | NMR structure of Ydj1 J-domain, a cytosolic Hsp40 from Saccharomyces cerevisiae | | Descriptor: | Yeast dnaJ protein 1 | | Authors: | Ciesielski, S.J, Tonelli, M, Lee, W, Cornilescu, G, Markley, J.L, Schilke, B.A, Ziegelhoffer, T, Craig, E.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Broadening the functionality of a J-protein/Hsp70 molecular chaperone system.
PLoS Genet., 13, 2017
|
|
