1F22
 
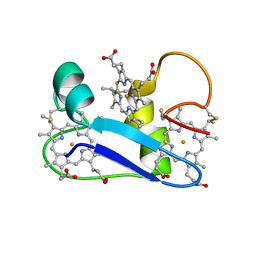 | | A PROTON-NMR INVESTIGATION OF THE FULLY REDUCED CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS. COMPARISON BETWEEN THE REDUCED AND THE OXIDIZED FORMS. | | Descriptor: | CYTOCHROME C7, HEME C | | Authors: | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Giudici-Orticoni, M.T. | | Deposit date: | 2000-05-23 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | A proton-NMR investigation of the fully reduced cytochrome c7 from Desulfuromonas acetoxidans. Comparison between the reduced and the oxidized forms.
Eur.J.Biochem., 266, 1999
|
|
1F6A
 
 | | Structure of the human ige-fc bound to its high affinity receptor fc(epsilon)ri(alpha) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR ALPHA-SUBUNIT, ... | | Authors: | Garman, S.C, Wurzburg, B.A, Tarchevskaya, S.S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2000-06-20 | | Release date: | 2000-07-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc (epsilon) RI (alpha).
Nature, 406, 2000
|
|
1F5Y
 
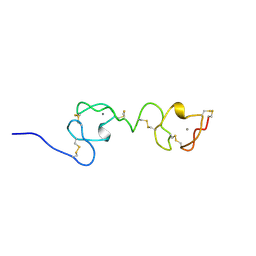 | | NMR STRUCTURE OF A CONCATEMER OF THE FIRST AND SECOND LIGAND-BINDING MODULES OF THE HUMAN LDL RECEPTOR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Atkins, A.R, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2000-06-18 | | Release date: | 2000-08-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a concatemer of the first and second ligand-binding modules of the human low-density lipoprotein receptor.
Protein Sci., 9, 2000
|
|
1FCY
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE RARBETA/GAMMA-SELECTIVE RETINOID CD564 | | Descriptor: | 6-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTALENE-2-CARBONYL)-NAPHTALENE-2-CARBOXYLIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1IXU
 
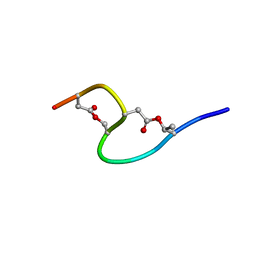 | | Solution structure of marinostatin, a protease inhibitor, containing two ester linkages | | Descriptor: | marinostatin | | Authors: | Kanaori, K, Kamei, K, Koyama, T, Yasui, T, Takano, R, Imada, C, Tajima, K, Hara, S. | | Deposit date: | 2002-07-04 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of marinostatin, a natural ester-linked protein protease inhibitor
Biochemistry, 44, 2005
|
|
1IHH
 
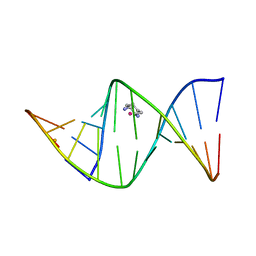 | | 2.4 ANGSTROM CRYSTAL STRUCTURE OF AN OXALIPLATIN 1,2-D(GPG) INTRASTRAND CROSS-LINK IN A DNA DODECAMER DUPLEX | | Descriptor: | 1R,2R-DIAMINOCYCLOHEXANE, 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', ... | | Authors: | Spingler, B, Whittington, D.A, Lippard, S.J. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 A crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex.
Inorg.Chem., 40, 2001
|
|
1FPI
 
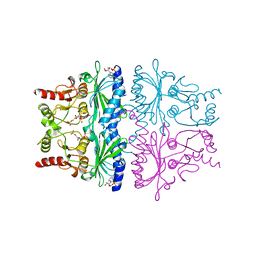 | | FRUCTOSE-1,6-BISPHOSPHATASE (D-FRUCTOSE-1,6-BISPHOSPHATE 1-PHOSPHOHYDROLASE) COMPLEXED WITH AMP, 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND POTASSIUM IONS (100 MM) | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Lipscomb, W.N. | | Deposit date: | 1995-06-02 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic evidence for the action of potassium, thallium, and lithium ions on fructose-1,6-bisphosphatase.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1FBZ
 
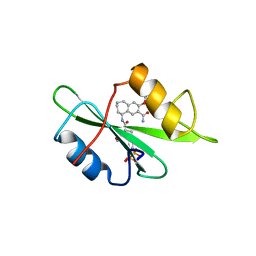 | | Structure-based design of a novel, osteoclast-selective, nonpeptide Src SH2 inhibitor with in vivo anti-resorptive activity | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK, {4-[2-ACETYLAMINO-2-(3-CARBAMOYL-2-CYCLOHEXYLMETHOXY-6,7,8,9-TETRAHYDRO-5H-BENZOCYCLOHEPTEN-5YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENYL}-PHOSPHONIC ACID | | Authors: | Shakespeare, W, Yang, M, Bohacek, R, Cerasoli, F, Stebbis, K, Sundaramoorthi, R, Vu, C, Pradeepan, S, Metcalf, C, Haraldson, C, Merry, T, Dalgarno, D, Narula, S, Hatada, M, Lu, X, Van Schravendijk, M.R, Adams, S, Violette, S, Smith, J, Guan, W, Bartlett, C, Herson, J, Iuliucci, J, Weigele, M, Sawyer, T. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of an osteoclast-selective, nonpeptide src homology 2 inhibitor with in vivo antiresorptive activity.
Proc.Natl.Acad.Sci.Usa, 97, 2000
|
|
1FCL
 
 | |
1FPJ
 
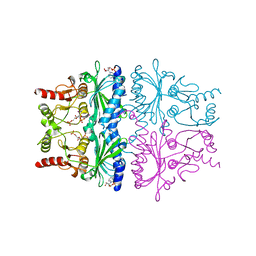 | | FRUCTOSE-1,6-BISPHOSPHATASE (D-FRUCTOSE-1,6-BISPHOSPHATE 1-PHOSPHOHYDROLASE) COMPLEXED WITH AMP, 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE, THALLIUM (10 MM) AND LITHIUM IONS (10 MM) | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Lipscomb, W.N. | | Deposit date: | 1995-06-02 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic evidence for the action of potassium, thallium, and lithium ions on fructose-1,6-bisphosphatase.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1JCP
 
 | | Solution structure of the lactam analogue EDap of HIV gp41 600-612 loop. | | Descriptor: | Edap : ACE-Ile-Trp-Glu-Ser-Gly-Lys-Leu-Ile-Dap-Thr-Thr-Ala ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-06-11 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1JC8
 
 | | Solution structure of lactam analogue (DDap) of gp41 600-612 loop of HIV | | Descriptor: | DDap: (ACE)IWGDSGKLI(DNP)TTA ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-06-08 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1FCZ
 
 | | ISOTYPE SELECTIVITY OF THE HUMAN RETINOIC ACID NUCLEAR RECEPTOR HRAR: THE COMPLEX WITH THE PANAGONIST RETINOID BMS181156 | | Descriptor: | 4-[3-OXO-3-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-PROPENYL]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-1 | | Authors: | Klaholz, B.P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-07-19 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis for isotype selectivity of the human retinoic acid nuclear receptor.
J.Mol.Biol., 302, 2000
|
|
1K2Y
 
 | |
1K35
 
 | |
1FP5
 
 | |
1FPU
 
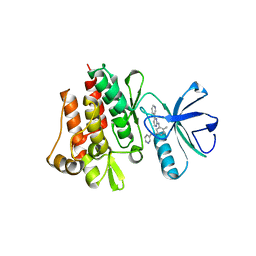 | | CRYSTAL STRUCTURE OF ABL KINASE DOMAIN IN COMPLEX WITH A SMALL MOLECULE INHIBITOR | | Descriptor: | N-[4-METHYL-3-[[4-(3-PYRIDINYL)-2-PYRIMIDINYL]AMINO]PHENYL]-3-PYRIDINECARBOXAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL | | Authors: | Schindler, T, Bornmann, W, Pellicena, P, Miller, W.T, Clarkson, B, Kuriyan, J. | | Deposit date: | 2000-08-31 | | Release date: | 2000-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism for STI-571 inhibition of abelson tyrosine kinase.
Science, 289, 2000
|
|
1FE1
 
 | | CRYSTAL STRUCTURE PHOTOSYSTEM II | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, CADMIUM ION, CHLOROPHYLL A, ... | | Authors: | Zouni, A, Witt, H.-T, Kern, J, Fromme, P, Krauss, N, Saenger, W, Orth, P. | | Deposit date: | 2000-07-20 | | Release date: | 2001-02-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution.
Nature, 409, 2001
|
|
1FM2
 
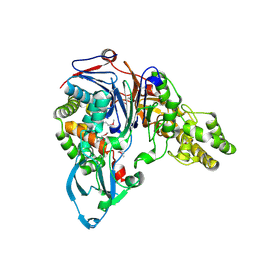 | | THE 2 ANGSTROM CRYSTAL STRUCTURE OF CEPHALOSPORIN ACYLASE | | Descriptor: | GLUTARYL 7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Kim, Y, Yoon, K.H, Khang, Y, Turley, S, Hol, W.G.J. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-15 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of cephalosporin acylase.
Structure Fold.Des., 8, 2000
|
|
1FQ3
 
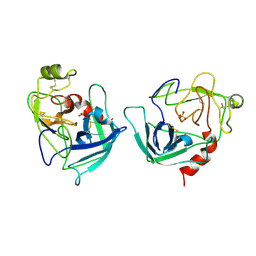 | | CRYSTAL STRUCTURE OF HUMAN GRANZYME B | | Descriptor: | GRANZYME B, SULFATE ION, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Estebanez-Perpina, E, Fuentes-Prior, P, Belorgey, D, Rubin, H, Bode, W. | | Deposit date: | 2000-09-03 | | Release date: | 2001-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the caspase activator human granzyme B, a proteinase highly specific for an Asp-P1 residue.
Biol.Chem., 381, 2000
|
|
1JOH
 
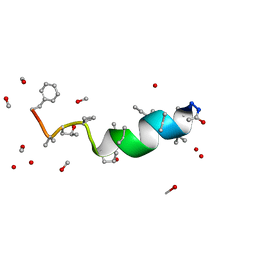 | |
1FGI
 
 | |
1JQN
 
 | |
1K25
 
 | |
1K1J
 
 | | BOVINE TRYPSIN-INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, N-ALPHA-(2-NAPHTHYLSULFONYL)-N(3-AMIDINO-L-PHENYLALANINYL)ISOPIPECOLINIC ACID METHYL ESTER, SULFATE ION, ... | | Authors: | Stubbs, M.T. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
J.Mol.Biol., 313, 2001
|
|
