7Y2P
 
 | |
7Q9Q
 
 | |
9B3C
 
 | | type 2 KD-mxyl filament of miniature tau macrocycle derived from 4R tauopathic fold | | Descriptor: | 1,3-dimethylbenzene, AMINO GROUP, GLY-SER-VAL-GLN-ILE-VAL-TYR, ... | | Authors: | Xu, X, Angera, J.I, Rajewski, H.B, Jiang, W, Del Valle, R.J. | | Deposit date: | 2024-03-18 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure-based design of seed-competent proteomimetic
macrocycles derived from 4R tauopathic folds
To Be Published
|
|
480D
 
 | |
5FXC
 
 | | Crystal structure of glycopeptide 22 in complex with scFv-SM3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCOPEPTIDE, SCFV-SM3 | | Authors: | Rojas-Ocariz, V, Companon, I, Aydillo, C, Castro-Lopez, J, Jimenez-Barbero, J, Hurtado-Guerrero, R, Avenoza, A, Zurbano, M.M, Corzana, F, Busto, J.H, Peregrina, J.M. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design of Alpha-S-Glycopeptides Derived from Muc1 with a Flexible and Solvent Exposed Sugar Moiety
J.Org.Chem., 81, 2016
|
|
4YCJ
 
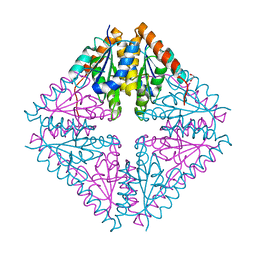 | | Structure of Acetobacter aceti PurE Y154F | | Descriptor: | 1,2-ETHANEDIOL, N5-carboxyaminoimidazole ribonucleotide mutase, SULFATE ION | | Authors: | Kappock, T.J, Sullivan, K.L. | | Deposit date: | 2015-02-20 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structure of a single tryptophan mutant of Acetobacter aceti PurE
To Be Published
|
|
6E60
 
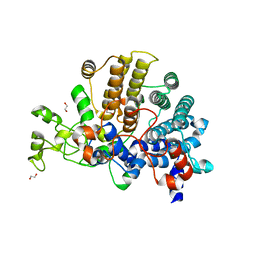 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-A | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, mixed-linkage glucan utilization locus (MLGUL) SGBP-B | | Authors: | Tamura, K, Gardill, B.R, Brumer, H, Van Petegem, F. | | Deposit date: | 2018-07-23 | | Release date: | 2019-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
9O0V
 
 | | Crystal structure of CRAF/MEK1 complex with PLX4720, CH5126766, and AMPPNP | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, N-(3-fluoro-4-{[4-methyl-2-oxo-7-(pyrimidin-2-yloxy)-2H-chromen-3-yl]methyl}pyridin-2-yl)-N'-methylsulfuric diamide, N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}propane-1-sulfonamide, ... | | Authors: | Jang, D.M, Eck, M.J. | | Deposit date: | 2025-04-03 | | Release date: | 2025-10-15 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Characterization and inhibitor sensitivity of ARAF, BRAF, and CRAF kinases.
J.Biol.Chem., 2025
|
|
9QFQ
 
 | | Cryo-EM structure of the cofilactin barbed end bound by AIP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2025-03-12 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Choreography of rapid actin filament disassembly by coronin, cofilin, and AIP1.
Cell, 2025
|
|
9Q1N
 
 | |
9OIO
 
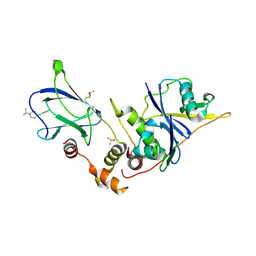 | | The von Hippel Lindau-ElonginB-ElonginC (VCB) complex with fragments 9 and 14 | | Descriptor: | 1-[(2-fluorophenyl)methyl]-4-(propan-2-yl)piperazine, 1-[6-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]-2,3-dihydroindol-1-yl]ethanone, DIMETHYL SULFOXIDE, ... | | Authors: | Amporndanai, K, Katinas, J.M, Chopra, A, Fesik, S.W. | | Deposit date: | 2025-05-06 | | Release date: | 2025-07-09 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NMR-Based Fragment Screen of the von Hippel-Lindau Elongin C&B Complex.
Acs Med.Chem.Lett., 16, 2025
|
|
9SFG
 
 | | Crystal structure of NLRP3 in complex with inhibitor NP3-742 | | Descriptor: | 5-methyl-~{N}-[(3~{R})-1-methylpiperidin-3-yl]-6-(2-methyl-1~{H}-pyrrolo[2,3-b]pyridin-6-yl)pyridazin-3-amine, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Srinivas, H. | | Deposit date: | 2025-08-19 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of NP3-742: A Structurally Diverse NLRP3 Inhibitor Identified through an Unusual Phenol Replacement.
J.Med.Chem., 2025
|
|
9UNS
 
 | | Apg mutant enzyme D448N of acarbose hydrolase from human gut flora K. grimontii TD1,complex with acarbose | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-5-[[(1~{S},4~{R},5~{S},6~{S})-3-(hydroxymethyl)-4,5,6-tris(oxidanyl)cyclohex-2-en-1-yl]amino]-6-methyl-oxane-2,3,4-triol-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, Maltodextrin glucosidase | | Authors: | Zhou, J.H, Huang, J.Y. | | Deposit date: | 2025-04-24 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular insights of acarbose metabolization catalyzed by acarbose-preferred glucosidase.
Nat Commun, 16, 2025
|
|
9US5
 
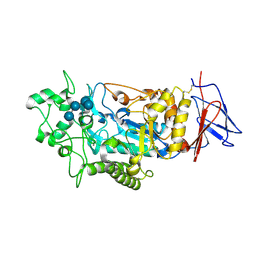 | |
9OG5
 
 | | SARS-COV-2-6P-MUT7 S PROTEIN-DY-III-281 complex 1 RBD up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Chandravanshi, M, Niu, L, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2025-04-30 | | Release date: | 2025-10-01 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of VE607 to generate analogs with improved neutralization activities against SARS-CoV-2 variants.
J.Virol., 2025
|
|
9Q2D
 
 | | Cryo-EM structure of ternary complex Ikaros-ZF2:CC-885:CRBN:DDB1 (molecular glue degrader) | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)urea, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | Authors: | Zhu, J, Pagarigan, B.E, Tran, E.T. | | Deposit date: | 2025-08-15 | | Release date: | 2025-10-01 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Application of Weighted Interaction-Fingerprints for Rationalizing Neosubstrate Potency and Selectivity of Cereblon-Based Molecular Glues.
J.Med.Chem., 68, 2025
|
|
9Q03
 
 | | Cryo-EM structure of ternary complex BCL6-CRBN-DDB1 with BCL6-760 (LDD, local refined) | | Descriptor: | (3R)-3-(6-{4-[(5-chloro-4-{[3-(3-hydroxy-3-methylbutyl)-1-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-5-yl]amino}pyrimidin-2-yl)(methyl)amino]piperidin-1-yl}-1-methyl-1H-indazol-3-yl)piperidine-2,6-dione, B-cell lymphoma 6 protein, Protein cereblon, ... | | Authors: | Zhu, J, Fang, W, Pagarigan, B. | | Deposit date: | 2025-08-12 | | Release date: | 2025-10-15 | | Last modified: | 2025-11-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Discovery of Potent and Selective BCL6 Ligand-Directed Degrader (LDD), BCL6-760.
J.Med.Chem., 68, 2025
|
|
9FD3
 
 | | CysG(N-16)-D190N mutant in complex with SAH from Kitasatospora cystarginea | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-16 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
7N4T
 
 | | Low conductance mechanosensitive channel YnaI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Low conductance mechanosensitive channel YnaI | | Authors: | Catalano, C, Ben-Hail, D, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structure of Mechanosensitive Channel YnaI Using SMA2000: Challenges and Opportunities.
Membranes (Basel), 11, 2021
|
|
9O0U
 
 | | Crystal structure of CRAF/MEK1 complex with PLX4720 and CH5126766 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, N-(3-fluoro-4-{[4-methyl-2-oxo-7-(pyrimidin-2-yloxy)-2H-chromen-3-yl]methyl}pyridin-2-yl)-N'-methylsulfuric diamide, N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}propane-1-sulfonamide, ... | | Authors: | Jang, D.M, Eck, M.J. | | Deposit date: | 2025-04-03 | | Release date: | 2025-10-15 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Characterization and inhibitor sensitivity of ARAF, BRAF, and CRAF kinases.
J.Biol.Chem., 2025
|
|
9VSQ
 
 | | The TUG-891-bound structure of TMEM175 | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, CHOLESTEROL, Endosomal/lysosomal proton channel TMEM175, ... | | Authors: | Zhu, X, Liu, H, Yin, W. | | Deposit date: | 2025-07-09 | | Release date: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural insights into the activation of TMEM175 by small molecule.
Neuron, 2025
|
|
9QFW
 
 | | Cryo-EM structure of the cofilactin barbed end bound by two AIP1 molecules | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2025-03-12 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Choreography of rapid actin filament disassembly by coronin, cofilin, and AIP1.
Cell, 2025
|
|
6UZF
 
 | | Crystal structure of the unliganded bromodomain of human BRD9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, DIMETHYL SULFOXIDE, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
9VSR
 
 | | The DCY1040 and TUG-891-bound structure of TMEM175 | | Descriptor: | (2S)-2-butyl-2-cyclohexyl-6,7-bis(fluoranyl)-5-[3-(1H-1,2,3,4-tetrazol-5-yl)propoxy]-3H-inden-1-one, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, CHOLESTEROL, ... | | Authors: | Zhu, X, Liu, H, Yin, W. | | Deposit date: | 2025-07-09 | | Release date: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural insights into the activation of TMEM175 by small molecule.
Neuron, 2025
|
|
8CHU
 
 | |
