2JFG
 
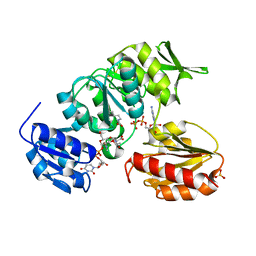 | | Crystal structure of MurD ligase in complex with UMA and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE, ... | | Authors: | Kotnik, M, Humljan, J, Contreras-Martel, C, Oblak, M, Kristan, K, Herve, M, Blanot, D, Urleb, U, Gobec, S, Dessen, A, Solmajer, T. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and functional characterization of enantiomeric glutamic acid derivatives as potential transition state analogue inhibitors of MurD ligase.
J. Mol. Biol., 370, 2007
|
|
2JFH
 
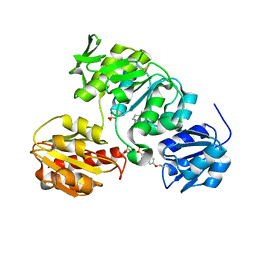 | | Crystal structure of MurD ligase in complex with L-Glu containing sulfonamide inhibitor | | Descriptor: | N-[(6-BUTOXYNAPHTHALEN-2-YL)SULFONYL]-L-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYL-ALANINE-D-GLUTAMATE LIGASE | | Authors: | Kotnik, M, Humljan, J, Contreras-Martel, C, Oblak, M, Kristan, K, Herve, M, Blanot, D, Urleb, U, Gobec, S, Dessen, A, Solmajer, T. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Characterization of Enantiomeric Glutamic Acid Derivatives as Potential Transition State Analogue Inhibitors of Murd Ligase.
J.Mol.Biol., 370, 2007
|
|
3EFR
 
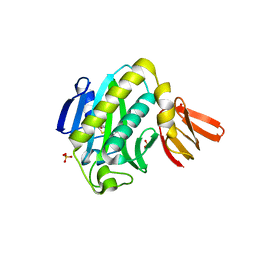 | | Biotin protein ligase R40G mutant from Aquifex aeolicus in complex with biotin | | Descriptor: | BIOTIN, Biotin [acetyl-CoA-carboxylase] ligase, SULFATE ION | | Authors: | Tron, C.M, McNae, I.W, Walkinshaw, M.D, Baxter, R.L, Campopiano, D.J. | | Deposit date: | 2008-09-10 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional studies of the biotin protein ligase from Aquifex aeolicus reveal a critical role for a conserved residue in target specificity.
J.Mol.Biol., 387, 2009
|
|
1WQ7
 
 | |
7B5N
 
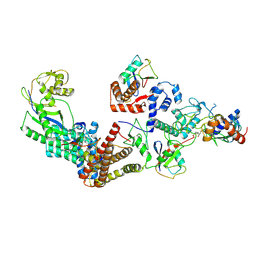 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: NEDD8-CUL1-RBX1-UBE2L3~Ub~ARIH1. | | Descriptor: | 5-azanylpentan-2-one, Cullin-1, E3 ubiquitin-protein ligase ARIH1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5S
 
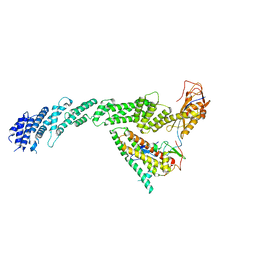 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-ARIH1 Ariadne. Transition State 1 | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase ARIH1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-07 | | Release date: | 2021-02-10 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5L
 
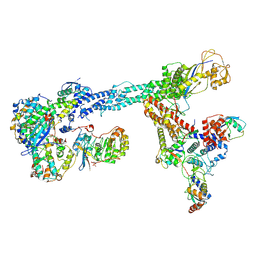 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: NEDD8-CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27-UBE2L3~Ub~ARIH1. Transition State 1 | | Descriptor: | 5-azanylpentan-2-one, Cullin-1, Cyclin-A2, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5M
 
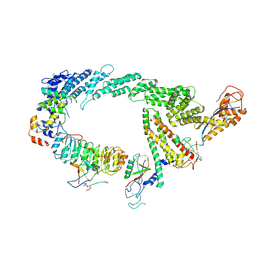 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-SKP1-SKP2-CKSHS1-p27~Ub~ARIH1. Transition State 2 | | Descriptor: | Cullin-1, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-17 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
1WPY
 
 | |
3EFS
 
 | | Biotin protein ligase from Aquifex aeolicus in complex with biotin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BIOTIN, Biotin [acetyl-CoA-carboxylase] ligase, ... | | Authors: | Tron, C.M, McNae, I.W, Walkinshaw, M.D, Baxter, R.L, Campopiano, D.J. | | Deposit date: | 2008-09-10 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional studies of the biotin protein ligase from Aquifex aeolicus reveal a critical role for a conserved residue in target specificity.
J.Mol.Biol., 387, 2009
|
|
2JFF
 
 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-[(6-BUTOXYNAPHTHALEN-2-YL)SULFONYL]-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Kotnik, M, Humljan, J, Contreras-Martel, C, Oblak, M, Kristan, K, Herve, M, Blanot, D, Urleb, U, Gobec, S, Dessen, A, Solmajer, T. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and Functional Characterization of Enantiomeric Glutamic Acid Derivatives as Potential Transition State Analogue Inhibitors of Murd Ligase.
J.Mol.Biol., 370, 2007
|
|
3LWB
 
 | | Crystal Structure of apo D-alanine:D-alanine Ligase (Ddl) from Mycobacterium tuberculosis | | Descriptor: | D-alanine--D-alanine ligase, NITRATE ION | | Authors: | Bruning, J.B, Murillo, A.C, Chacon, O, Barletta, R.G, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Mycobacterium tuberculosis D-Alanine:D-Alanine Ligase, a Target of the Antituberculosis Drug D-Cycloserine.
Antimicrob.Agents Chemother., 55, 2011
|
|
4D1C
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH bromovinylhydantoin bound. | | Descriptor: | (5Z)-5-[(3-bromophenyl)methylidene]imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1D
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | Descriptor: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1B
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | Descriptor: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1A
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | Descriptor: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
5H7S
 
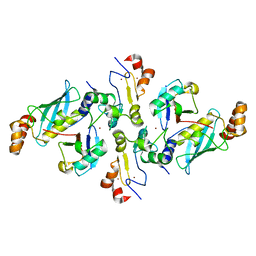 | |
3QVG
 
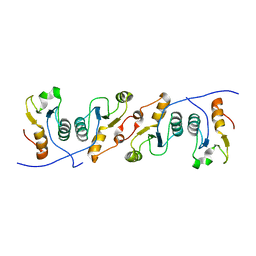 | | XRCC1 bound to DNA ligase | | Descriptor: | DNA ligase 3, DNA repair protein XRCC1 | | Authors: | Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes.
Nucleic Acids Res., 39, 2011
|
|
1P8L
 
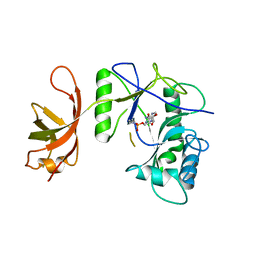 | | New Crystal Structure of Chlorella Virus DNA Ligase-Adenylate | | Descriptor: | ADENOSINE MONOPHOSPHATE, PBCV-1 DNA ligase | | Authors: | Odell, M, Malinina, L, Teplova, M, Shuman, S. | | Deposit date: | 2003-05-07 | | Release date: | 2003-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Analysis of the DNA Joining Repertoire of Chlorella Virus DNA ligase and a New Crystal Structure of the Ligase-Adenylate Intermediate
Nucleic Acids Res., 31, 2003
|
|
1JH5
 
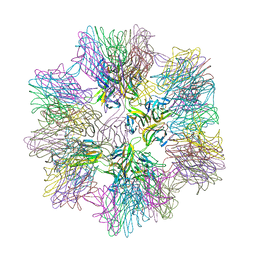 | | Crystal Structure of sTALL-1 of TNF family ligand | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 13B | | Authors: | Liu, Y, Xu, L, Opalka, N, Shu, H.-B, Zhang, G. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of sTALL-1 reveals a virus-like assembly of TNF family ligands.
Cell(Cambridge,Mass.), 108, 2002
|
|
3PC8
 
 | | X-ray crystal structure of the heterodimeric complex of XRCC1 and DNA ligase III-alpha BRCT domains. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA ligase 3, DNA repair protein XRCC1, ... | | Authors: | Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2010-10-21 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The structural basis for partitioning of the XRCC1/DNA ligase III-{alpha} BRCT-mediated dimer complexes.
Nucleic Acids Res., 39, 2011
|
|
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | Descriptor: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7R
 
 | | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide | | Descriptor: | 5-azanyl-1-oxidanyl-pentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
8R5H
 
 | | Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 5-azanylpentan-2-one, Bromodomain-containing protein 4, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
1IOV
 
 | | COMPLEX OF D-ALA:D-ALA LIGASE WITH ADP AND A PHOSPHORYL PHOSPHONATE | | Descriptor: | 2-[(1-AMINO-ETHYL)-PHOSPHATE-PHOSPHINOYLOXY]-BUTYRIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALA:D-ALA LIGASE, ... | | Authors: | Knox, J.R, Moews, P.C, Fan, C. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D-alanine:D-alanine ligase: phosphonate and phosphinate intermediates with wild type and the Y216F mutant.
Biochemistry, 36, 1997
|
|
