6UVK
 
 | | OXA-48 bound by inhibitor CDD-97 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-(2-ethoxyphenyl)piperazin-1-yl]-1,3,5-triazin-2-yl}piperidine-4-carboxylic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T.G. | | Deposit date: | 2019-11-02 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying Oxacillinase-48 Carbapenemase Inhibitors Using DNA-Encoded Chemical Libraries.
Acs Infect Dis., 6, 2020
|
|
3W3M
 
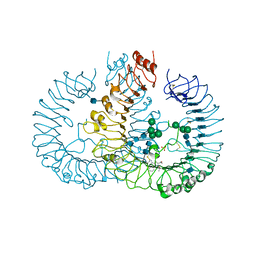 | | Crystal structure of human TLR8 in complex with Resiquimod (R848) crystal form 2 | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
4GN5
 
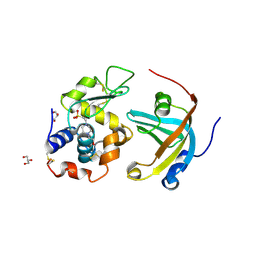 | | OBody AM3L15 bound to hen egg-white lysozyme | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Steemson, J.D, Liddament, M.T. | | Deposit date: | 2012-08-16 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Tracking Molecular Recognition at the Atomic Level with a New Protein Scaffold Based on the OB-Fold.
Plos One, 9, 2014
|
|
8X0C
 
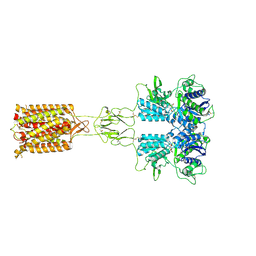 | | Human FL Metabotropic glutamate receptor 5, mGlu5-5M with quisqualate and VU0424465, conformer 1 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Vinothkumar, K.R, Lebon, G, Cannone, G. | | Deposit date: | 2023-11-04 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational diversity in class C GPCR positive allosteric modulation.
Nat Commun, 16, 2025
|
|
7UMB
 
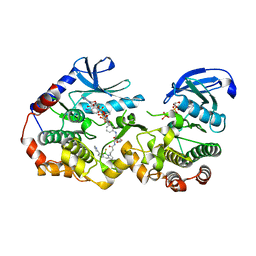 | | NanoBRET tracer Tram-bo bound to a KSR2-MEK1 complex | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Kinase suppressor of Ras 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Marsiglia, W.M, Khan, K.M, Dar, A.C. | | Deposit date: | 2022-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.231 Å) | | Cite: | Live-cell target engagement of allosteric MEKi on MEK-RAF/KSR-14-3-3 complexes.
Nat.Chem.Biol., 20, 2024
|
|
4UI1
 
 | | Crystal structure of the human RGMC-BMP2 complex | | Descriptor: | 1,2-ETHANEDIOL, BONE MORPHOGENETIC PROTEIN 2, CHLORIDE ION, ... | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3BEP
 
 | | Structure of a sliding clamp on DNA | | Descriptor: | 1-(3-hydroxypropyl)-2-{(1E,3E,5E)-5-[1-(3-hydroxypropyl)-3,3-dimethyl-1,3-dihydro-2H-indol-2-ylidene]penta-1,3-dien-1-y l}-3,3-dimethyl-3H-indolium, DNA (5'-D(*DTP*DTP*DTP*DTP*DAP*DTP*DAP*DCP*DGP*DAP*DTP*DGP*DGP*DG)-3'), DNA (5'-D(P*DCP*DCP*DCP*DAP*DTP*DCP*DGP*DTP*DAP*DT)-3'), ... | | Authors: | Georgescu, R.E, Kim, S.S, Yurieva, O, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of a sliding clamp on DNA
Cell(Cambridge,Mass.), 132, 2008
|
|
9N3S
 
 | | Crystal structure of the fungal mannosyltransferase Och1 reveals active site primed for N-glycan binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Initiation-specific alpha-1,6-mannosyltransferase, ... | | Authors: | Kelly, E.T.R, Rodionov, D, Berghuis, A.M. | | Deposit date: | 2025-01-31 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the fungal mannosyltransferase Och1 reveals active site primed for N-glycan binding.
Plos One, 20, 2025
|
|
4ZEB
 
 | | PBP AccA from A. tumefaciens C58 in complex with agrocinopine A | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
5GLM
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compost microbial metagenome in complex with xylotriose, calcium-free form. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Glycoside hydrolase family 43, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
4UJ6
 
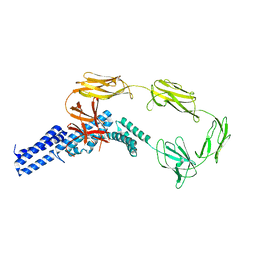 | | Structure of surface layer protein SbsC, domains 1-6 | | Descriptor: | SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of Surface Layer Protein Sbsc, Domains 1-6
To be Published
|
|
8X4S
 
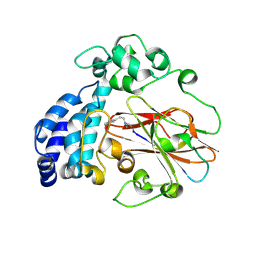 | | The L-tryptophan specific decarboxylase PsiD covalent bonding with tryptamine | | Descriptor: | (2~{R})-2-[2-(1~{H}-indol-3-yl)ethylamino]propanoic acid, L-tryptophan decarboxylase | | Authors: | Meng, C.Y, Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2023-11-15 | | Release date: | 2024-11-20 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for psilocybin biosynthesis.
Nat Commun, 16, 2025
|
|
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
6I24
 
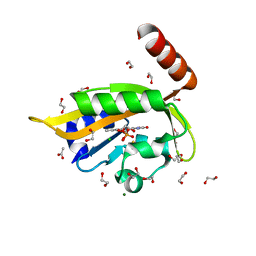 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, ACETATE ION, ... | | Authors: | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | Deposit date: | 2018-10-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
9F0I
 
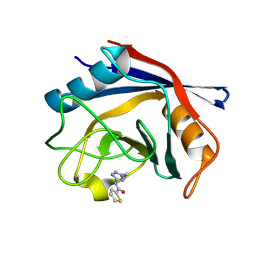 | | Human Cyclophilin D in complex with fragment | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ~{N}-(2-ethyl-1,2,3,4-tetrazol-5-yl)thiophene-2-carboxamide | | Authors: | Silva, D.O, Graedler, U, Bandeiras, T.M. | | Deposit date: | 2024-04-16 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human Cyclophilin D in complex with fragment
To Be Published
|
|
4ZFL
 
 | |
7Q0F
 
 | | Structure of Candida albicans 80S ribosome in complex with phyllanthoside | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
9F0Q
 
 | | VIM-2 in complex with GKV53 (5d) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Bartels, K, Prester, A, Bosman, R, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
9F0P
 
 | | VIM-2 in complex with GKV61 (5c) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Bartels, K, Prester, A, Bosman, R, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
2L9D
 
 | | Solution structure of the protein YP_546394.1, the first structural representative of the pfam family PF12112 | | Descriptor: | Uncharacterized protein | | Authors: | Mohanty, B, Serrano, P, Geralt, M, Horst, R, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-02-08 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the protein YP_546394.1, the first structural representative of the pfam family PF12112
To be Published
|
|
6IBS
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 6 | | Descriptor: | CALCIUM ION, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
4UOY
 
 | | Crystal structure of YgjG in complex with Pyridoxal-5'-phosphate | | Descriptor: | FORMIC ACID, GLYCEROL, PUTRESCINE AMINOTRANSFERASE, ... | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2014-06-11 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Structure of Putrescine Aminotransferase from Escherichia Coli Provides Insights Into the Substrate Specificity Among Class III Aminotransferases.
Plos One, 9, 2014
|
|
7VUH
 
 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUJ
 
 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
6IBV
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd 1 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
