4OR7
 
 | |
4IC2
 
 | | Crystal structure of the XIAP RING domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, NICKEL (II) ION, ZINC ION | | Authors: | Linke, K, Nakatani, Y, Day, C.L. | | Deposit date: | 2012-12-09 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of ubiquitin transfer by XIAP, a dimeric RING E3 ligase
Biochem.J., 450, 2013
|
|
4O7J
 
 | |
4IBP
 
 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target EFI-900011, with bound glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase-like protein YibF, SULFATE ION | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens Pf-5, target IFI-900011, with bound glutathione
To be Published
|
|
4OSV
 
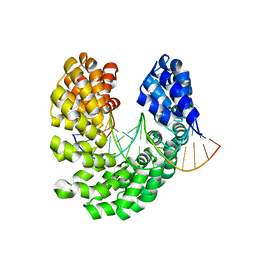 | | Crystal structure of the S505M mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
2XZV
 
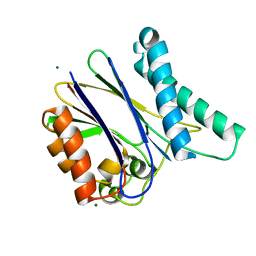 | |
2A7C
 
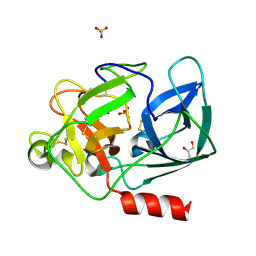 | | On the Routine Use of Soft X-Rays in Macromolecular Crystallography, Part III- The Optimal Data Collection Wavelength | | Descriptor: | Elastase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Mueller-Dieckmann, C, Panjikar, S, Tucker, P.A, Weiss, M.S. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part III. The optimal data-collection wavelength.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4OTT
 
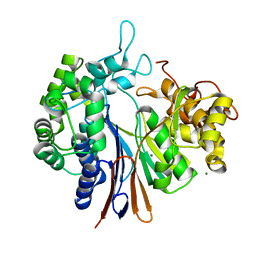 | | Crystal structure of the gamma-glutamyltranspeptidase from Bacillus licheniformis. | | Descriptor: | Gamma glutamyl transpeptidase, Gamma-glutamyltranspeptidase, MAGNESIUM ION | | Authors: | Merlino, A. | | Deposit date: | 2014-02-14 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Low resolution X-ray structure of gamma-glutamyltranspeptidase from Bacillus licheniformis: Opened active site cleft and a cluster of acid residues potentially involved in the recognition of a metal ion.
Biochim.Biophys.Acta, 1844, 2014
|
|
4IGJ
 
 | | Crystal structure of Maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, target EFI-507175 | | Descriptor: | Maleylacetoacetate isomerase, SULFATE ION | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, target EFI-507175
To be Published
|
|
3ZSC
 
 | | Catalytic function and substrate recognition of the pectate lyase from Thermotoga maritima | | Descriptor: | 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, GLYCEROL, PECTATE TRISACCHARIDE-LYASE, ... | | Authors: | McDonough, M.A, Thymark, M, Frisner, H, Hotchkiss, A, Sonksen, C, Bjornvad, M, Johansen, K.S, Larsen, S. | | Deposit date: | 2011-06-24 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Catalytic Function and Substrate Recognition of the Pectate Lyase from Thermotoga Maritima
To be Published
|
|
4IKT
 
 | | Crystal structure of truncated (delta 1-89) human methionine aminopeptidase Type 1 in complex with N1-(5-chloro-6-methyl-2-(pyridin-2-yl)pyrimidin-4-yl)-N2-(5-(trifluoromethyl)pyridin-2-yl)ethane-1,2-diamine | | Descriptor: | COBALT (II) ION, GLYCEROL, Methionine aminopeptidase 1, ... | | Authors: | Addlagatta, A, Kishor, C, Arya, T. | | Deposit date: | 2012-12-28 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification, Biochemical and Structural Evaluation of Species-Specific Inhibitors against Type I Methionine Aminopeptidases
J.Med.Chem., 56, 2013
|
|
4OVX
 
 | | Crystal structure of Xylose isomerase domain protein from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, Xylose isomerase domain protein TIM barrel | | Authors: | Chang, C, Bigelow, L, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-22 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of Xylose isomerase domain protein from Planctomyces limnophilus DSM 3776
To be published
|
|
1T6B
 
 | | Crystal structure of B. anthracis Protective Antigen complexed with human Anthrax toxin receptor | | Descriptor: | Anthrax toxin receptor 2, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Santelli, E, Bankston, L.A, Leppla, S.H, Liddington, R.C. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a complex between anthrax toxin and its host cell receptor
Nature, 430, 2004
|
|
2A9M
 
 | |
5RGD
 
 | | Crystal Structure of Kemp Eliminase HG3.14 with bound transition state analogue, 277K | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.14, SULFATE ION | | Authors: | Broom, A, Rakotoharisoa, R.V, Thompson, M.C, Fraser, J.S, Chica, R.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico.
Nat Commun, 11, 2020
|
|
5RHJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z126932614 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-[(methylsulfonyl)methyl]-1H-benzimidazole, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SXQ
 
 | |
4IHK
 
 | | Crystal structure of the Collagen VI alpha3 N5 domain R1061Q | | Descriptor: | Collagen alpha3(VI) | | Authors: | Mikolajek, H, Becker, A.K.A, Paulsson, M, Wagener, R, Werner, J.M. | | Deposit date: | 2012-12-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A structure of a collagen VI VWA domain displays N and C termini at opposite sides of the protein
Structure, 22, 2014
|
|
1T9S
 
 | | Catalytic Domain Of Human Phosphodiesterase 5A in Complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-18 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
4IEW
 
 | |
1TBB
 
 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With Rolipram | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, ROLIPRAM, ... | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
1TEJ
 
 | | Crystal structure of a disintegrin heterodimer at 1.9 A resolution. | | Descriptor: | disintegrin chain A, disintegrin chain B | | Authors: | Bilgrami, S, Kaur, P, Yadav, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Disintegrin Heterodimer from Saw-Scaled Viper (Echis carinatus) at 1.9 A Resolution
Biochemistry, 44, 2005
|
|
4IFS
 
 | | Crystal structure of the hSSRP1 Middle domain | | Descriptor: | CHLORIDE ION, FACT complex subunit SSRP1 | | Authors: | Zhang, W.J, Zeng, F.X, Shao, C, Liu, Y.W, Niu, L.W, Li, X, Teng, M.K. | | Deposit date: | 2012-12-15 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the hSSRP1 Middle domain
To be Published
|
|
1TG4
 
 | | Design of specific inhibitors of groupII phospholipase A2(PLA2): Crystal structure of the complex formed between russells viper PLA2 and designed peptide Phe-Leu-Ala-Tyr-Lys at 1.7A resolution | | Descriptor: | FLAYK peptide, Phospholipase A2, SULFATE ION | | Authors: | Singh, N, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Ethayathulla, A.S, Singh, T.P. | | Deposit date: | 2004-05-28 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of specific inhibitors of groupII phospholipase A2(PLA2): Crystal structure of the complex formed between russells viper PLA2 and designed peptide Phe-Leu-Ala-Tyr-Lys at 1.7A resolution
TO BE PUBLISHED
|
|
2ABA
 
 | | Structure of reduced PETN reductase in complex with progesterone | | Descriptor: | FLAVIN MONONUCLEOTIDE, ISOPROPYL ALCOHOL, PROGESTERONE, ... | | Authors: | Khan, H, Barna, T, Bruce, N.C, Munro, A.W, Leys, D, Scrutton, N.S. | | Deposit date: | 2005-07-15 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Proton transfer in the oxidative half-reaction of pentaerythritol tetranitrate reductase
Febs J., 272, 2005
|
|
