5JXG
 
 | | Structure of the unliganded form of the proprotein convertase furin. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2CKY
 
 | | Structure of the Arabidopsis thaliana thiamine pyrophosphate riboswitch with its regulatory ligand | | Descriptor: | MAGNESIUM ION, NUCLEIC ACID, OSMIUM ION, ... | | Authors: | Thore, S, Leibundgut, M, Ban, N. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Eukaryotic Thiamine Pyrophosphate Riboswitch with its Regulatory Ligand.
Science, 312, 2006
|
|
7UPN
 
 | |
6SJ7
 
 | |
2GPV
 
 | | Estrogen Related Receptor-gamma ligand binding domain complexed with 4-hydroxy-tamoxifen and a SMRT peptide | | Descriptor: | 4-HYDROXYTAMOXIFEN, Estrogen-related receptor gamma, Nuclear receptor corepressor 2 | | Authors: | Wang, L, Zuercher, W.J, Consler, T.G, Lambert, M.H, Miller, A.B, Osband-miller, L.A, McKee, D.D, Willson, T.M, Nolte, R.T. | | Deposit date: | 2006-04-18 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation.
J.Biol.Chem., 281, 2006
|
|
2XQ5
 
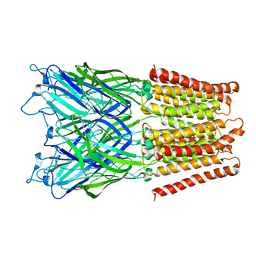 | | Pentameric ligand gated ion channel GLIC in complex with tetraethylarsonium (TEAs) | | Descriptor: | ARSENIC, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQ7
 
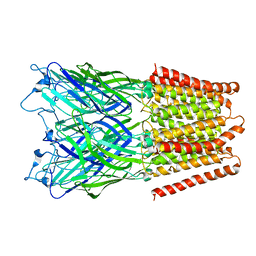 | | Pentameric ligand gated ion channel GLIC in complex with cadmium ion (Cd2+) | | Descriptor: | CADMIUM ION, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2I83
 
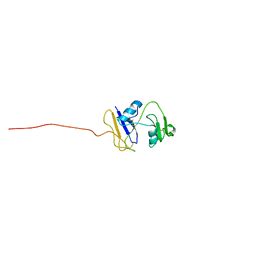 | | hyaluronan-binding domain of CD44 in its ligand-bound form | | Descriptor: | CD44 antigen | | Authors: | Takeda, M, Ogino, S, Umemoto, R, Sakakura, M, Kajiwara, M, Sugahara, K.N, Hayasaka, H, Miyasaka, M, Terasawa, H, Shimada, I. | | Deposit date: | 2006-09-01 | | Release date: | 2006-11-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Ligand-induced Structural Changes of the CD44 Hyaluronan-binding Domain Revealed by NMR
J.Biol.Chem., 281, 2006
|
|
3R75
 
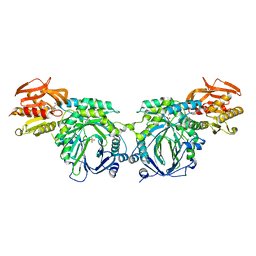 | | Crystal structure of 2-amino-2-desoxyisochorismate synthase (ADIC) synthase PhzE from Burkholderia lata 383 in complex with benzoate, pyruvate, glutamine and contaminating Zn2+ | | Descriptor: | Anthranilate/para-aminobenzoate synthases component I, BENZOIC ACID, MAGNESIUM ION, ... | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
2GPO
 
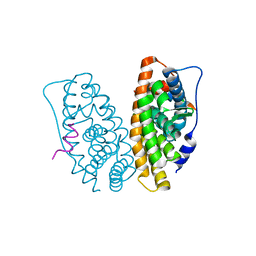 | | Estrogen Related Receptor-gamma ligand binding domain complexed with a synthetic peptide from RIP140 | | Descriptor: | Estrogen-related receptor gamma, Nuclear receptor-interacting protein 1 | | Authors: | Wang, L, Zuercher, W.J, Consler, T.G, Lambert, M.H, Miller, A.B, Osband-miller, L.A, McKee, D.D, Willson, T.M, Nolte, R.T. | | Deposit date: | 2006-04-18 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation.
J.Biol.Chem., 281, 2006
|
|
2GPU
 
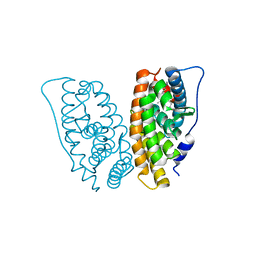 | | Estrogen Related Receptor-gamma ligand binding domain complexed with 4-hydroxy-tamoxifen | | Descriptor: | 4-HYDROXYTAMOXIFEN, Estrogen-related receptor gamma | | Authors: | Wang, L, Zuercher, W.J, Consler, T.G, Lambert, M.H, Miller, A.B, Osband-Miller, L.A, McKee, D.D, Willson, T.M, Nolte, R.T. | | Deposit date: | 2006-04-18 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation.
J.Biol.Chem., 281, 2006
|
|
4L48
 
 | |
1P22
 
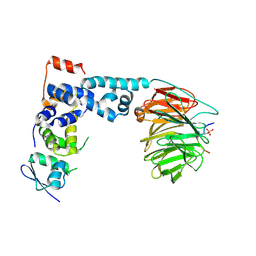 | | Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity on the SCFbeta-TrCP1 ubiquitin ligase | | Descriptor: | Beta-catenin, F-box/WD-repeat protein 1A, Skp1 | | Authors: | Wu, G, Xu, G, Schulman, B.A, Jeffrey, P.D, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a beta-TrCP1-Skp1-beta-Catenin complex: destruction motif binding and lysine specificity of the SCFbeta-TrCP1 ubiquitin ligase
Mol.Cell, 11, 2003
|
|
3R77
 
 | | Crystal structure of the D38A mutant of isochorismatase PhzD from Pseudomonas fluorescens 2-79 in complex with 2-amino-2-desoxyisochorismate ADIC | | Descriptor: | (5S,6S)-6-amino-5-[(1-carboxyethenyl)oxy]cyclohexa-1,3-diene-1-carboxylic acid, CHLORIDE ION, Probable isochorismatase | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
2GP7
 
 | | Estrogen Related Receptor-gamma ligand binding domain | | Descriptor: | CALCIUM ION, Estrogen-related receptor gamma | | Authors: | Wang, L, Zuercher, W.J, Consler, T.G, Lambert, M.H, Miller, A.B, Osband-miller, L.A, McKee, D.D, Willson, T.M, Nolte, R.T. | | Deposit date: | 2006-04-17 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation.
J.Biol.Chem., 281, 2006
|
|
1YJE
 
 | | Crystal structure of the rNGFI-B ligand-binding domain | | Descriptor: | Orphan nuclear receptor NR4A1 | | Authors: | Flaig, R, Greschik, H, Peluso-Iltis, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain.
J.Biol.Chem., 280, 2005
|
|
1T97
 
 | |
1F8Z
 
 | | NMR STRUCTURE OF THE SIXTH LIGAND-BINDING MODULE OF THE LDL RECEPTOR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Clayton, D.J, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2000-07-05 | | Release date: | 2000-10-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional NMR structure of the sixth ligand-binding module of the human LDL receptor: comparison of two adjacent modules with different ligand binding specificities.
FEBS Lett., 479, 2000
|
|
2ORX
 
 | | Structural Basis for Ligand Binding and Heparin Mediated Activation of Neuropilin | | Descriptor: | Neuropilin-1 | | Authors: | Vander Kooi, C.W, Jusino, M.A, Perman, B, Neau, D.B, Bellamy, H.D, Leahy, D.J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ligand and heparin binding to neuropilin B domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ORZ
 
 | | Structural Basis for Ligand Binding and Heparin Mediated Activation of Neuropilin | | Descriptor: | Neuropilin-1, Tuftsin | | Authors: | Vander Kooi, C.W, Jusino, M.A, Perman, B, Neau, D.B, Bellamy, H.D, Leahy, D.J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for ligand and heparin binding to neuropilin B domains.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8U2Q
 
 | |
8U2P
 
 | |
6IJR
 
 | | Human PPARgamma ligand binding domain complexed with SB1495 | | Descriptor: | 16 mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1S,2R)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
7KE0
 
 | |
3GKK
 
 | |
