7TCG
 
 | | BceAB nucleotide-free conformation | | Descriptor: | 4-amino-4-deoxy-1-O-[(S)-hydroxy{[(2E,6E,10Z,14Z,18Z,22E,26E,30E)-3,7,11,15,19,23,27,31,35-nonamethylhexatriaconta-2,6,10,14,18,22,26,30,34-nonaen-1-yl]oxy}phosphoryl]-alpha-L-arabinopyranose, Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2021-12-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational snapshots of the bacitracin sensing and resistance transporter BceAB.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TCH
 
 | | BceAB E169Q variant ATP-bound conformation | | Descriptor: | 4-amino-4-deoxy-1-O-[(S)-hydroxy{[(2E,6E,10Z,14Z,18Z,22E,26E,30E)-3,7,11,15,19,23,27,31,35-nonamethylhexatriaconta-2,6,10,14,18,22,26,30,34-nonaen-1-yl]oxy}phosphoryl]-alpha-L-arabinopyranose, ADENOSINE-5'-TRIPHOSPHATE, Bacitracin export ATP-binding protein BceA, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2021-12-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational snapshots of the bacitracin sensing and resistance transporter BceAB.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8G4C
 
 | | BceABS ATPgS high res TM | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G4D
 
 | | BceABS ATPgS tilted BceS | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G3F
 
 | | BceAB-S nucleotide free BceS state 1 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G3L
 
 | | BceAB-S nucleotide free BceS state 2 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-08 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G3B
 
 | | BceAB-S nucleotide free TM state 2 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G3A
 
 | | BceAB-S nucleotide free TM state 1 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
4FG2
 
 | |
4FG4
 
 | |
4FER
 
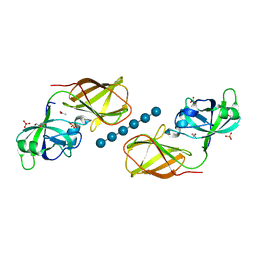 | | Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with cellohexaose | | Descriptor: | ACETIC ACID, Expansin-yoaJ, GLYCEROL, ... | | Authors: | Georgelis, N, Yennawar, N.H, Cosgrove, D.J. | | Deposit date: | 2012-05-30 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FFT
 
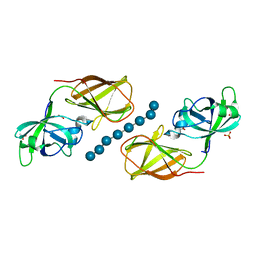 | | Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with mixed-linkage glucan | | Descriptor: | ACETIC ACID, Expansin-yoaJ, SULFATE ION, ... | | Authors: | Georgelis, N, Yennawar, N.H, Cosgrove, D.J. | | Deposit date: | 2012-06-01 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UFM
 
 | |
3UF7
 
 | |
3UDG
 
 | | Structure of Deinococcus radiodurans SSB bound to ssDNA | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', Single-stranded DNA-binding protein, THYMIDINE-5'-PHOSPHATE | | Authors: | George, N.P, Ngo, K.V, Chitteni-Patu, S, Norais, C.A, Battista, J.R, Cox, M.M, Keck, J.L. | | Deposit date: | 2011-10-28 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Cellular Dynamics of Deinococcus radiodurans Single-stranded DNA (ssDNA)-binding Protein (SSB)-DNA Complexes.
J.Biol.Chem., 287, 2012
|
|
6UVO
 
 | | Structure of antibody 3G12 bound to the central conserved domain of RSV G | | Descriptor: | 3G12 Fab Heavy chain, 3G12 Fab light chain, Major surface glycoprotein G | | Authors: | Fedechkin, S.O, George, N.L, Nunez Castrejon, A.M, Dillen, J, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2019-11-03 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformational Flexibility in Respiratory Syncytial Virus G Neutralizing Epitopes.
J.Virol., 94, 2020
|
|
5WNA
 
 | | Structure of antibody 3D3 bound to the central conserved region of RSV G | | Descriptor: | Major surface glycoprotein G, mAb 3D3 Fab heavy chain, mAb 3D3 Fab light chain | | Authors: | Fedechkin, S.O, George, N.L, Wolff, J.T, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of respiratory syncytial virus G antigen bound to broadly neutralizing antibodies.
Sci Immunol, 3, 2018
|
|
5WN9
 
 | | Structure of antibody 2D10 bound to the central conserved region of RSV G | | Descriptor: | Major surface glycoprotein G, scFv 2D10 | | Authors: | Fedechkin, S.O, George, N.L, Wolff, J.T, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structures of respiratory syncytial virus G antigen bound to broadly neutralizing antibodies.
Sci Immunol, 3, 2018
|
|
5WNB
 
 | | Structure of antibody 3D3 bound to the linear epitope of RSV G | | Descriptor: | ACETIC ACID, Major surface glycoprotein G, ZINC ION, ... | | Authors: | Fedechkin, S.O, George, N.L, Wolff, J.T, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of respiratory syncytial virus G antigen bound to broadly neutralizing antibodies.
Sci Immunol, 3, 2018
|
|
4L48
 
 | |
4NL4
 
 | | PriA Helicase Bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Primosome assembly protein PriA, ZINC ION | | Authors: | Bhattacharyya, B, George, N.P, Keck, J.L. | | Deposit date: | 2013-11-13 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural mechanisms of PriA-mediated DNA replication restart.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NL8
 
 | | PriA Helicase Bound to SSB C-terminal Tail Peptide | | Descriptor: | Primosome assembly protein PriA, Single-stranded DNA-binding protein, ZINC ION | | Authors: | Bhattacharyya, B, George, N.P, Thurmes, T.M, Keck, J.L. | | Deposit date: | 2013-11-13 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.08 Å) | | Cite: | Structural mechanisms of PriA-mediated DNA replication restart.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4JS7
 
 | |
4JJO
 
 | |
4JCW
 
 | |
