3LVA
 
 | |
4NO9
 
 | | yCP in complex with Z-Leu-Leu-Leu-epoxyketone | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]-L-leucinamide, N-[(benzyloxy)carbonyl]-L-leucyl-N-{(1R,2S)-1-hydroxy-4-methyl-1-[(2R)-2-methyloxiran-2-yl]pentan-2-yl}-L-leucinamide, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3LW5
 
 | | Improved model of plant photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3g54890, BETA-CAROTENE, ... | | Authors: | Nelson, N, Toporik, H. | | Deposit date: | 2010-02-23 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure determination and improved model of plant photosystem I
J.Biol.Chem., 285, 2010
|
|
3M2U
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-08 | | Release date: | 2010-09-15 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
4NE1
 
 | | Human MHF1 MHF2 DNA complexes | | Descriptor: | Centromere protein S, Centromere protein X, DNA (26-MER) | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
4NIA
 
 | | Satellite Tobacco Mosaic Virus Refined at room temperature to 1.8 A Resolution using NCS Restraints | | Descriptor: | Coat protein, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Satellite tobacco mosaic virus refined to 1.4 angstrom resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3MIW
 
 | | Crystal Structure of Rotavirus NSP4 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural glycoprotein 4 | | Authors: | Chacko, A.R, Read, R.J, Dodson, E.J, Rao, D.C, Suguna, K. | | Deposit date: | 2010-04-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new pentameric structure of rotavirus NSP4 revealed by molecular replacement.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4NCO
 
 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP gp120, BG505 SOSIP gp41, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Lyumkis, D, Ward, A.B, Wilson, I.A. | | Deposit date: | 2013-10-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Crystal structure of a soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
4NO6
 
 | | yCP in complex with Z-Leu-Leu-Leu-vinylsulfone | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(3S)-5-methyl-1-(methylsulfonyl)hexan-3-yl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3MG6
 
 | | Structure of yeast 20S open-gate proteasome with Compound 6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N~2~-{[(1S)-6-methoxy-3-oxo-2,3-dihydro-1H-inden-1-yl]acetyl}-N-{(1S)-1-[(4-methylbenzyl)carbamoyl]-3-phenylpropyl}-L-threoninamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
4NRT
 
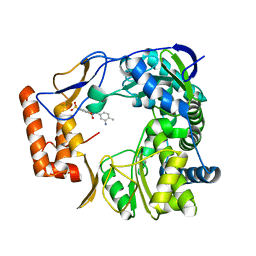 | | Human Norovirus polymerase bound to Compound 6 (suramin derivative) | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, hNV-RdRp | | Authors: | Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Milani, M, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
4NO1
 
 | | yCP in complex with Z-Leu-Leu-Leu-B(OH)2 | | Descriptor: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(1R)-1-(dihydroxyboranyl)-3-methylbutyl]-L-leucinamide, Probable proteasome subunit alpha type-7, ... | | Authors: | Stein, M.L, Cui, H, Beck, P, Dubiella, C, Voss, C, Krueger, A, Schmidt, B, Groll, M. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the alpha-Ketoamide Electrophile as an Auspicious Reversible Lead Motif.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3MEN
 
 | |
3MK7
 
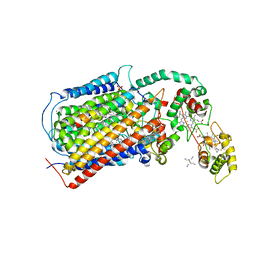 | | The structure of CBB3 cytochrome oxidase | | Descriptor: | 30-mer peptide, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Buschmann, S, Warkentin, E, Michel, H, Ermler, U. | | Deposit date: | 2010-04-14 | | Release date: | 2010-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of cbb3 Cytochrome Oxidase Provides Insights into Proton Pumping
Science, 329, 2010
|
|
3J7P
 
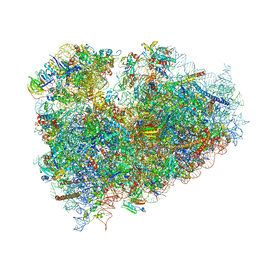 | | Structure of the 80S mammalian ribosome bound to eEF2 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
3JBP
 
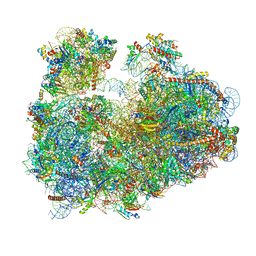 | | Cryo-electron microscopy reconstruction of the Plasmodium falciparum 80S ribosome bound to E-tRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein eS1, ... | | Authors: | Sun, M, Li, W, Blomqvist, K, Das, S, Hashem, Y, Dvorin, J.D, Frank, J. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Dynamical features of the Plasmodium falciparum ribosome during translation.
Nucleic Acids Res., 43, 2015
|
|
3JAM
 
 | | CryoEM structure of 40S-eIF1A-eIF1 complex from yeast | | Descriptor: | 18S rRNA, MAGNESIUM ION, RACK1, ... | | Authors: | Llacer, J.L, Hussain, T, Ramakrishnan, V. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex.
Mol.Cell, 59, 2015
|
|
3JB9
 
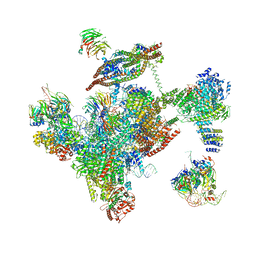 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
3JCJ
 
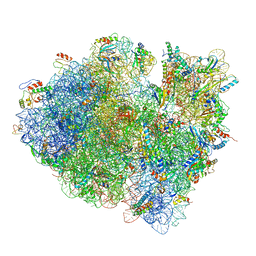 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2015-12-18 | | Release date: | 2016-03-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
3K0D
 
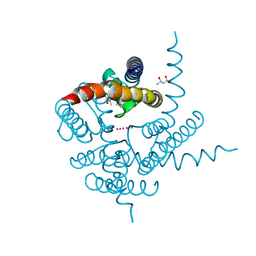 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, K+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein NaK | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4PEI
 
 | | Dbr1 in complex with synthetic branched RNA analog | | Descriptor: | GLYCEROL, NICKEL (II) ION, RNA (5'-R(*(G46)P*U)-3'), ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
4PJO
 
 | | Minimal U1 snRNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ETHANOL, ... | | Authors: | Kondo, Y, Oubridge, C, van Roon, A.M, Nagai, K. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5' splice site recognition.
Elife, 4, 2015
|
|
4POT
 
 | |
3K2N
 
 | |
3K35
 
 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
