8FU6
 
 | | GCGR-Gs complex in the presence of RAMP2 | | Descriptor: | Glucagon derivative ZP3780, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Krishna Kumar, K, O'Brien, E.S, Wang, H, Montabana, E, Kobilka, B.K. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Negative allosteric modulation of the glucagon receptor by RAMP2.
Cell, 186, 2023
|
|
6F23
 
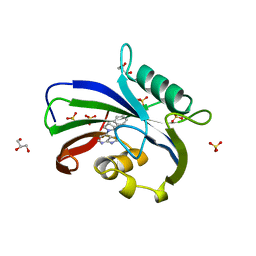 | | Complex between MTH1 and compound 16 (a 4-amino-7-azaindole derivative) | | Descriptor: | 4-[(2~{R})-2-phenylpyrrolidin-1-yl]-1~{H}-pyrrolo[2,3-b]pyridine, 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, ... | | Authors: | Viklund, J, Tresaugues, L, Talagas, A, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
8FO0
 
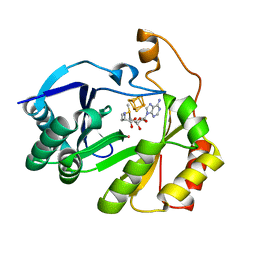 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to a partially cleaved SAM molecule | | Descriptor: | IRON/SULFUR CLUSTER, POTASSIUM ION, Pyruvate formate-lyase 1-activating enzyme, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FOL
 
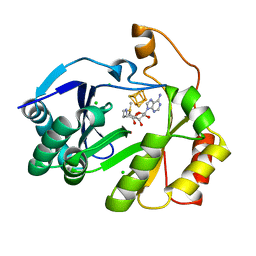 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM, alternate crystal form | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2022-12-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FSI
 
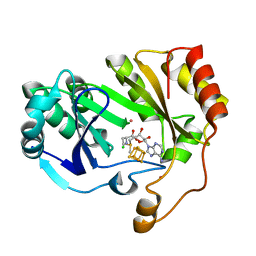 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | Authors: | Moody, J.D, Galambas, A, Lawrence, C.M, Broderick, J.B. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
5JVY
 
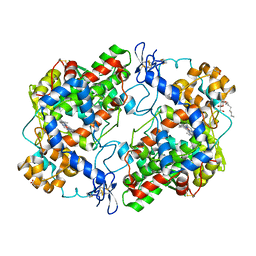 | | Crystal structure of S121P murine COX-2 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, ... | | Authors: | Orlando, B.J, Malkowski, M.G, Dong, L. | | Deposit date: | 2016-05-11 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Fatty Acid Binding to the Allosteric Subunit of Cyclooxygenase-2 Relieves a Tonic Inhibition of the Catalytic Subunit.
J.Biol.Chem., 291, 2016
|
|
7AKE
 
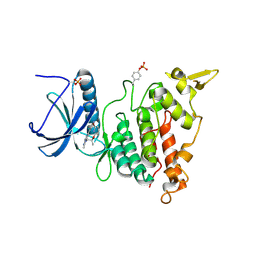 | | Structure of DYRK1A in complex with compound 58 | | Descriptor: | 4-[3-[(2~{S})-2-(6-bromanylpyridin-2-yl)oxypropyl]-2-methyl-imidazo[4,5-b]pyridin-5-yl]pyridine-2,6-diamine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AKB
 
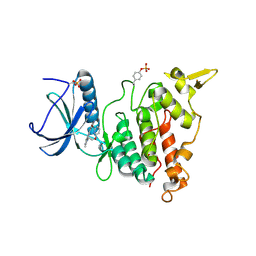 | | Structure of DYRK1A in complex with compound 56 | | Descriptor: | 4-[3-[2-(6-bromanylpyridin-2-yl)oxyethyl]-2-methyl-imidazo[4,5-b]pyridin-5-yl]pyridine-2,6-diamine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AJY
 
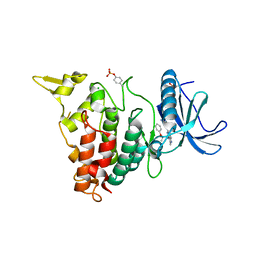 | | Structure of DYRK1A in complex with compound 51 | | Descriptor: | 4-[2-methyl-3-[(2~{R})-2-phenoxypropyl]imidazo[4,5-b]pyridin-5-yl]pyridine-2,6-diamine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AKA
 
 | | Structure of DYRK1A in complex with compound 54 | | Descriptor: | 4-[2-methyl-3-[(2~{R})-2-pyridin-2-yloxypropyl]imidazo[4,5-b]pyridin-5-yl]pyridine-2,6-diamine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AK2
 
 | | Structure of DYRK1A in complex with compound 53 | | Descriptor: | 4-[2-methyl-3-(2-pyridin-2-yloxyethyl)imidazo[4,5-b]pyridin-5-yl]pyridine-2,6-diamine, DIMETHYL SULFOXIDE, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ... | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
4YMJ
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-[6-(benzylamino)imidazo[1,2-b]pyridazin-3-yl]benzonitrile, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
6EYN
 
 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
5JE0
 
 | | Crystal structure of Burkholderia glumae ToxA with bound S-adenosylhomocysteine (SAH) and 1,6-didemethyltoxoflavin | | Descriptor: | Methyl transferase, S-ADENOSYL-L-HOMOCYSTEINE, pyrimido[5,4-e][1,2,4]triazine-5,7(6H,8H)-dione | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
8DI5
 
 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with VH domain F6 (focused refinement of RBD and VH F6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH F6 | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Subramaniam, S. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Potent and broad neutralization of SARS-CoV-2 variants of concern (VOCs) including omicron sub-lineages BA.1 and BA.2 by biparatopic human VH domains.
Iscience, 25, 2022
|
|
5IWM
 
 | | 2.5A structure of GSK945237 with S.aureus DNA gyrase and DNA. | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*TP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*AP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Miles, T.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8CRQ
 
 | | Local refinement of Band 3-I transmembrane domains, class 1 of erythrocyte ankyrin-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHOLESTEROL, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5KGP
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with chitosan | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-alpha-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
8CRR
 
 | | Local refinement of Band 3-III transmembrane domains, class 1 of erythrocyte ankyrin-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHOLESTEROL, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-11 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4YE2
 
 | |
7KDM
 
 | | Ricin bound to VHH antibody V5G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-RON nanobody, CHLORIDE ION, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7KC9
 
 | | Ricin bound to VHH antibody V5G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ricin chain A, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-05 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7KBI
 
 | | Ricin bound to VHH antibody V5E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ricin chain A, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-02 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.049 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
5JDY
 
 | | Crystal structure of Burkholderia glumae ToxA Y7F mutant with bound S-adenosylhomocysteine (SAH) and toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Methyl transferase, ... | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
5IIE
 
 | |
