7WL4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor SLP-50 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[2-ethyl-6-(4-methylpiperazin-1-yl)-3-oxidanylidene-2,7-diazatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaen-9-yl]-2,4-bis(fluoranyl)benzenesulfonamide | | Authors: | Zhang, C, Wang, C, Li, W, Zhang, Y, Xu, Y, Sun, L. | | Deposit date: | 2022-01-12 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design, synthesis, and anticancer evaluation of ammosamide B with pyrroloquinoline derivatives as novel BRD4 inhibitors.
Bioorg.Chem., 127, 2022
|
|
7WKY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13153 | | Descriptor: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
2MZU
 
 | |
6SJ7
 
 | |
2N5N
 
 | | Structure of an N-terminal domain of CHD4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Silva, A.P.G, Mackay, J.P. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminal Region of Chromodomain Helicase DNA-binding Protein 4 (CHD4) Is Essential for Activity and Contains a High Mobility Group (HMG) Box-like-domain That Can Bind Poly(ADP-ribose).
J.Biol.Chem., 291, 2016
|
|
6XA1
 
 | |
6XUZ
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 4 | | Descriptor: | 6-[1-[(2~{S})-1-methoxypropan-2-yl]-6-[(3~{S})-3-methylmorpholin-4-yl]imidazo[4,5-c]pyridin-2-yl]-3-methyl-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XV3
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 3 | | Descriptor: | 3-methyl-6-[6-[(3~{S})-3-methylmorpholin-4-yl]-1-[(1~{S})-1-phenylethyl]imidazo[4,5-c]pyridin-2-yl]-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SWN
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH iBET-BD1 (GSK778) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(methoxymethyl)-1-[(1~{R})-1-phenylethyl]-8-[[(3~{S})-pyrrolidin-3-yl]methoxy]imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4, ... | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
2MUU
 
 | |
8G16
 
 | |
8G15
 
 | |
8G17
 
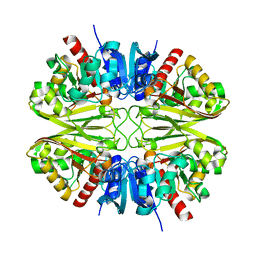 | | CryoEM structure of wild-type GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Choi, W.Y, Wu, H, Cheng, Y.F. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Efficient tagging of endogenous proteins in human cell lines for structural studies by single-particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4I8X
 
 | | Crystal structure of rabbit LDHA in complex with AP27460 | | Descriptor: | 6-phenylpyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
8G14
 
 | |
6XJT
 
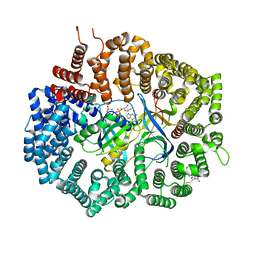 | | Crystal Structure of KPT-8602 bound to CRM1 (537-DLTVK-541 to GLCEQ) | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M, Chook, Y.M. | | Deposit date: | 2020-06-24 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Recurrent XPO1 mutations alter pathogenesis of chronic lymphocytic leukemia.
J Hematol Oncol, 14, 2021
|
|
8G13
 
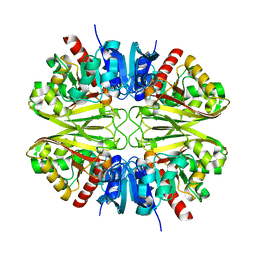 | |
8G12
 
 | |
2N0T
 
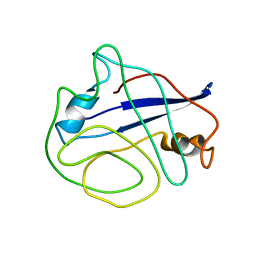 | | Structural ensemble of the enzyme cyclophilin reveals an orchestrated mode of action at atomic resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Chi, C.N, Voegeli, B, Bibow, S, Strotz, D, Orts, J, Guntert, P, Riek, R. | | Deposit date: | 2015-03-13 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Structural Ensemble for the Enzyme Cyclophilin Reveals an Orchestrated Mode of Action at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4HXS
 
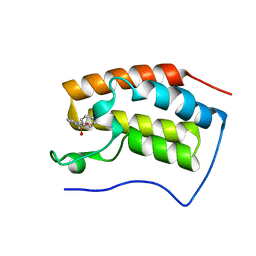 | | Brd4 Bromodomain 1 complex with N-[3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)PHENYL]-1-PHENYLMETHANESULFONAMIDE inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]-1-phenylmethanesulfonamide | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
5MEY
 
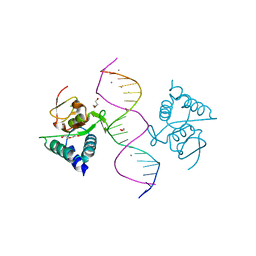 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
8FTA
 
 | |
2MS4
 
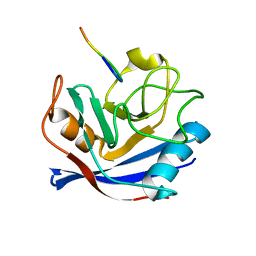 | | Cyclophilin a complexed with a fragment of crk-ii | | Descriptor: | Peptide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Jankowski, W, Saleh, T, Rossi, P, Kalodimos, C. | | Deposit date: | 2014-07-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cyclophilin A promotes cell migration via the Abl-Crk signaling pathway.
Nat.Chem.Biol., 12, 2016
|
|
6XEB
 
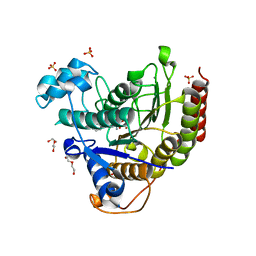 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH KETONE INHIBITOR (COMPOUND E) | | Descriptor: | 5-{(1S)-7,7-dihydroxy-1-[(1-methylazetidine-3-carbonyl)amino]nonyl}-2-phenyl-1H-imidazole-4-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Clausen, D. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of a selective HDAC inhibitor aimed at reactivating the HIV latent reservoir.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
2N3O
 
 | | Structure of PTB RRM1(41-163) bound to an RNA stemloop containing a structured loop derived from viral internal ribosomal entry site RNA | | Descriptor: | Polypyrimidine tract-binding protein 1, RNA (5'-R(*GP*GP*GP*AP*CP*CP*UP*GP*GP*UP*CP*UP*UP*UP*CP*CP*AP*GP*GP*UP*CP*CP*C)-3') | | Authors: | Maris, C, Jayne, S.F, Damberger, F.F, Ravindranathan, S, Allain, F.H.-T. | | Deposit date: | 2015-06-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | C-terminal helix folding upon pyrimidine-rich hairpin binding to PTB RRM1. Implications for PTB function in Encephalomyocarditis virus IRES activity.
To be Published
|
|
