5UBY
 
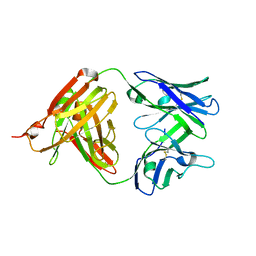 | | Fab structure of anti-HIV-1 gp120 mAb 1A8 | | 分子名称: | Heavy chain of Fab fragment of anti-HIV1 gp120 mAb 1A8, Light chain of Fab fragment of anti-HIV1 gp120 mAb 1A8 | | 著者 | Pan, R, Kong, X.-P. | | 登録日 | 2016-12-21 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | High Antibody Diversity and Low Inter-clonal Competition Favor Production of Functional Neutralizing Antibodies
To be Published
|
|
5UBZ
 
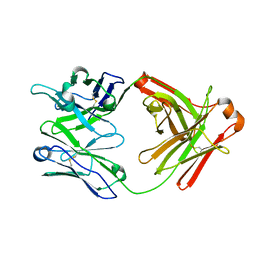 | | Fab structure of HIV gp120 specific mAb 1E12 | | 分子名称: | Anti-HIV1 gp120 mAb 1E12 Fab heavy chain, Anti-HIV1 gp120 mAb 1E12 Fab light chain | | 著者 | Pan, R, Kong, X.-P. | | 登録日 | 2016-12-21 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | High Antibody Diversity and Low Inter-clonal Competition Favor Production of Functional Neutralizing Antibodies
To be published
|
|
3TOE
 
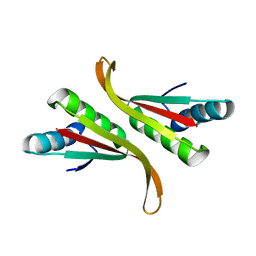 | | Structure of Mth10b | | 分子名称: | DNA/RNA-binding protein Alba | | 著者 | Pan, X.M, Zhang, N, Liu, Y.F, Liu, X. | | 登録日 | 2011-09-05 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.197 Å) | | 主引用文献 | Molecular mechanism underlying the interaction of typical Sac10b family proteins with DNA.
Plos One, 7, 2012
|
|
6VU2
 
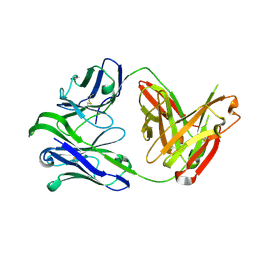 | | M1214_N1 Fab structure | | 分子名称: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | 著者 | Pan, R, Kong, X. | | 登録日 | 2020-02-14 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
3IVH
 
 | |
3IVI
 
 | | Design and Synthesis of Potent BACE-1 Inhibitors with Cellular Activity: Structure-Activity Relationship of P1 Substituents | | 分子名称: | Beta-secretase 1, GLYCEROL, N-[(1S,2R)-3-{[(5S)-5-(3-tert-butylphenyl)-4,5,6,7-tetrahydro-1H-indazol-5-yl]amino}-1-(3,5-difluorobenzyl)-2-hydroxypropyl]acetamide, ... | | 著者 | Pan, H. | | 登録日 | 2009-09-01 | | 公開日 | 2010-01-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Design and synthesis of cell potent BACE-1 inhibitors: structure-activity relationship of P1' substituents.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3K1R
 
 | |
5BTU
 
 | |
5BU3
 
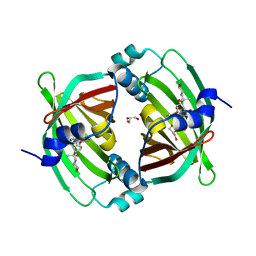 | | Crystal Structure of Diels-Alderase PyrI4 in complex with its product | | 分子名称: | (4S,4aS,6aS,8R,9R,10aR,13R,14aS,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,5,6,6a,7,8,9,10,10a,13,14,18a,18b-hexadecahydro-1H-14a,17-(metheno)benzo[b]naphtho[2,1-h]azacyclododecine-16,18(15H,17H)-dione, GLYCEROL, PyrI4 | | 著者 | Pan, L, Guo, Y, Liu, J. | | 登録日 | 2015-06-03 | | 公開日 | 2016-02-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.897 Å) | | 主引用文献 | Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4
Cell Chem Biol, 23, 2016
|
|
5XGV
 
 | |
5YT6
 
 | |
5ZFL
 
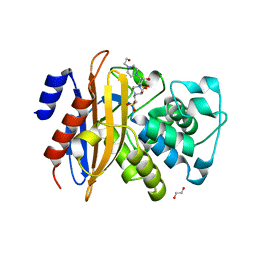 | | Crystal structure of beta-lactamase PenP mutant E166Y | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase | | 著者 | Pan, X, Zhao, Y. | | 登録日 | 2018-03-06 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The hydrolytic water molecule of Class A beta-lactamase relies on the acyl-enzyme intermediate ES* for proper coordination and catalysis.
Sci Rep, 10, 2020
|
|
5ZG6
 
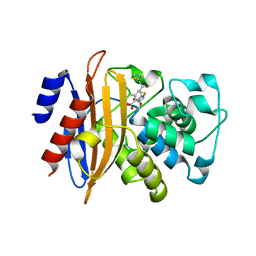 | |
5ZFT
 
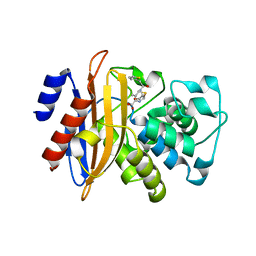 | |
5F5N
 
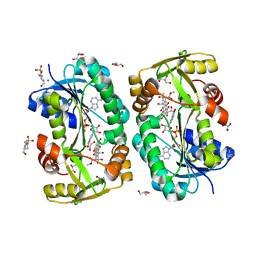 | |
5F5L
 
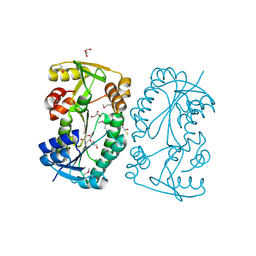 | |
8EA2
 
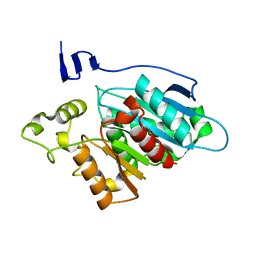 | |
8E83
 
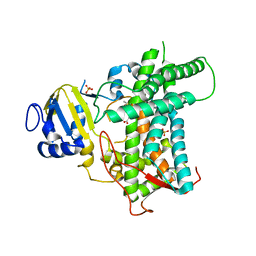 | |
8EA1
 
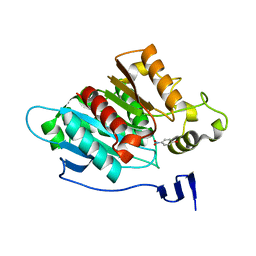 | |
7MSL
 
 | |
7SN0
 
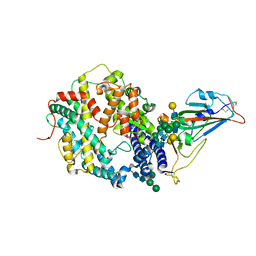 | | Crystal structure of spike protein receptor binding domain of escape mutant SARS-CoV-2 from immunocompromised patient (d146*) in complex with human receptor ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, J, Abraham, J, Clark, S. | | 登録日 | 2021-10-27 | | 公開日 | 2021-12-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN1
 
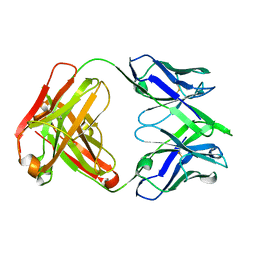 | |
7SN3
 
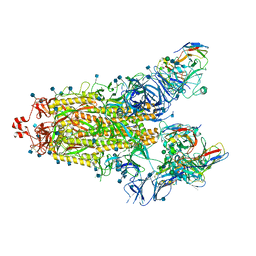 | | Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, J, Abraham, J, Shankar, S. | | 登録日 | 2021-10-27 | | 公開日 | 2021-12-08 | | 最終更新日 | 2022-02-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN2
 
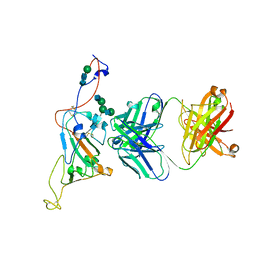 | | Structure of human SARS-CoV-2 neutralizing antibody C1C-A3 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pan, J, Abraham, J, Yang, P, Shankar, S. | | 登録日 | 2021-10-27 | | 公開日 | 2021-12-08 | | 最終更新日 | 2022-02-02 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
4I6H
 
 | |
