7YDL
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A/T268I/A184V/A82T in complex with N-imidazolyl-hexanoyl-L-phenylalanine
To Be Published
|
|
3KST
 
 | |
4MOG
 
 | | Pyranose 2-oxidase V546C mutant with 3-fluorinated glucose | | Descriptor: | 3-deoxy-3-fluoro-beta-D-glucopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
3KVC
 
 | | Crystal structure of bovine RPE65 at 1.9 angstrom resolution | | Descriptor: | FE (II) ION, Retinoid isomerohydrolase | | Authors: | Kiser, P.D, Lodowski, D.T, Golczak, M, Palczewski, K. | | Deposit date: | 2009-11-29 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of membrane structural integrity for RPE65 retinoid isomerization activity.
J.Biol.Chem., 285, 2010
|
|
3KT7
 
 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
2QG4
 
 | | Crystal structure of human UDP-glucose dehydrogenase product complex with UDP-glucuronate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kavanagh, K.L, Guo, K, Bunkoczi, G, Savitsky, P, Pilka, E, Bhatia, C, Niesen, F, Smee, C, Berridge, G, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
2QWX
 
 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
2GR2
 
 | |
3F1Q
 
 | | Human dihydroorotate dehydrogenase in complex with a leflunomide derivative inhibitor 1 | | Descriptor: | (2Z)-N-biphenyl-4-yl-2-cyano-3-hydroxybut-2-enamide, ACETIC ACID, Dihydroorotate dehydrogenase, ... | | Authors: | Heikkila, T, Davies, M, McConkey, G.A, Fishwick, C.W.G, Parsons, M.R, Johnson, A.P. | | Deposit date: | 2008-10-28 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases
J.Med.Chem., 52, 2009
|
|
2GR6
 
 | |
2GRU
 
 | | Crystal structure of 2-deoxy-scyllo-inosose synthase complexed with carbaglucose-6-phosphate, NAD+ and Co2+ | | Descriptor: | (1R,2S,3S,4R)-5-METHYLENECYCLOHEXANE-1,2,3,4-TETRAOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-deoxy-scyllo-inosose synthase, ... | | Authors: | Nango, E, Kumasaka, T. | | Deposit date: | 2006-04-25 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of 2-deoxy-scyllo-inosose synthase, a key enzyme in the biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics, in complex with a mechanism-based inhibitor and NAD+
Proteins, 70, 2008
|
|
2R1T
 
 | |
2GUW
 
 | |
4MOK
 
 | | Pyranose 2-oxidase H167A mutant soaked with 3-fluorinated galactose (not bound) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DODECAETHYLENE GLYCOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
2GUG
 
 | | NAD-dependent formate dehydrogenase from Pseudomonas sp.101 in complex with formate | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Formate dehydrogenase, ... | | Authors: | Filippova, E.V, Polyakov, K.M, Tikhonova, T.V, Boiko, K.M, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2006-04-30 | | Release date: | 2006-05-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the complex of NAD-dependent formate dehydrogenase from
metylotrophic bacterium Pseudomonas sp.101 with formate.
KRISTALLOGRAFIYA, 51, 2006
|
|
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | Descriptor: | CAFFEINE, Chitinase | | Authors: | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | Deposit date: | 2009-02-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
2QFX
 
 | |
3G77
 
 | | Bacterial cytosine deaminase V152A/F316C/D317G mutant | | Descriptor: | Cytosine deaminase, FE (III) ION | | Authors: | Stoddard, B, Zhao, L. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacterial cytosine deaminase mutants created by molecular engineering show improved 5-fluorocytosine-mediated cell killing in vitro and in vivo.
Cancer Res., 69, 2009
|
|
3G8Q
 
 | |
2QHA
 
 | |
3GA0
 
 | | CtBP1/BARS Gly172->Glu mutant structure: impairing NAD(H) binding and dimerization | | Descriptor: | C-terminal-binding protein 1, FORMIC ACID | | Authors: | Nardini, M, Valente, C, Ricagno, S, Luini, A, Corda, D, Bolognesi, M. | | Deposit date: | 2009-02-16 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | CtBP1/BARS Gly172-->Glu mutant structure: impairing NAD(H)-binding and dimerization
Biochem.Biophys.Res.Commun., 381, 2009
|
|
2GZ3
 
 | | Structure of Aspartate Semialdehyde Dehydrogenase (ASADH) from Streptococcus pneumoniae complexed with NADP and aspartate-semialdehyde | | Descriptor: | (2R)-2-AMINO-4-OXOBUTANOIC ACID, Aspartate beta-semialdehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Faehnle, C.R, Le Coq, J, Liu, X, Viola, R.E. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Examination of key intermediates in the catalytic cycle of aspartate-beta-semialdehyde dehydrogenase from a gram-positive infectious bacteria.
J.Biol.Chem., 281, 2006
|
|
2QV4
 
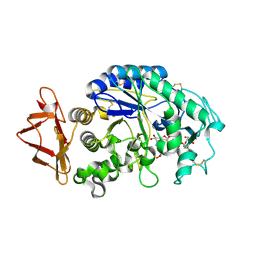 | | Human pancreatic alpha-amylase complexed with nitrite and acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5R,6S)-4-{[alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl]oxy}-5,6-dihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Williams, L.K, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-08-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
2H56
 
 | |
2QX4
 
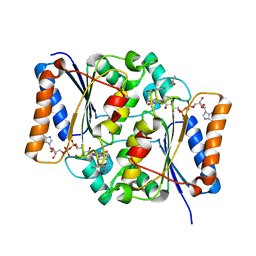 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
