1OQB
 
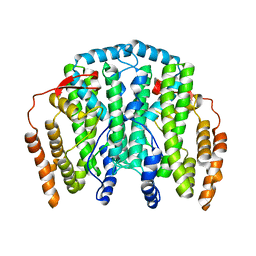 | | The Crystal Structure of the one-iron form of the di-iron center in Stearoyl Acyl Carrier Protein Desaturase from Ricinus Communis (Castor Bean). | | Descriptor: | Acyl-[acyl-carrier protein] desaturase, FE (II) ION | | Authors: | Moche, M, Shanklin, J, Ghoshal, A.K, Lindqvist, Y. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Azide and Acetate Complexes plus two iron-depleted Crystal Structures of the Di-iron Enzyme delta9 Stearoyl-ACP Desaturase-Implications for Oxygen Activation and Catalytic Intermediates
J.Biol.Chem., 278, 2003
|
|
5QND
 
 | |
4YVE
 
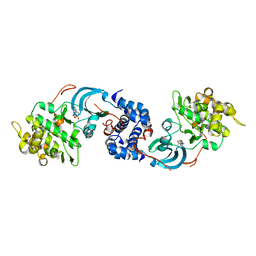 | | ROCK 1 bound to methoxyphenyl thiazole inhibitor | | Descriptor: | 2-(3-methoxyphenyl)-N-[4-(pyridin-4-yl)-1,3-thiazol-2-yl]acetamide, Rho-associated protein kinase 1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-03-19 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Pyridine-Based Rho Kinase (ROCK) Inhibitors.
J.Med.Chem., 58, 2015
|
|
2EZH
 
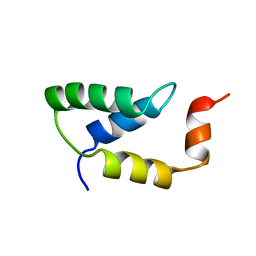 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZI
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, 30 STRUCTURES | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
4YDM
 
 | | High resolution crystal structure of human transthyretin bound to ligand and conjugates of 3-(5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl)phenyl fluorosulfate | | Descriptor: | 2,6-dichloro-4-[5-(3-hydroxyphenyl)-1,3,4-oxadiazol-2-yl]phenol, Transthyretin | | Authors: | Connelly, S, Bradbury, N.C. | | Deposit date: | 2015-02-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Fluorogenic Aryl Fluorosulfate for Intraorganellar Transthyretin Imaging in Living Cells and in Caenorhabditis elegans.
J.Am.Chem.Soc., 137, 2015
|
|
2BRX
 
 | |
4DSY
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment CC24201 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-phenylpyridine-3-carboxylic acid, ... | | Authors: | Feder, D, Hussein, W.M, Clayton, D.J, Kan, M, Schenk, G, McGeary, R.P, Guddat, L.W. | | Deposit date: | 2012-02-20 | | Release date: | 2012-09-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
7E7T
 
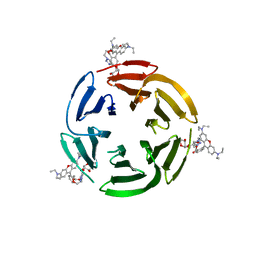 | | Crystal structure of RSL mutant in complex with sugar Ligand | | Descriptor: | 2-[2-[4-[[(2R,3S,4R,5S,6S)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-1,2,3-triazol-1-yl]ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, Fucose-binding lectin protein,Fucose-binding lectin protein,Fucose-binding lectin protein | | Authors: | Li, L, Chen, G.S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of RSL mutant-R17A/R108A/R199A in complex with R2F
To Be Published
|
|
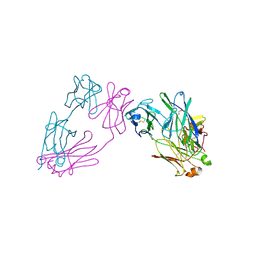
3MCG
 
 | |
7E7R
 
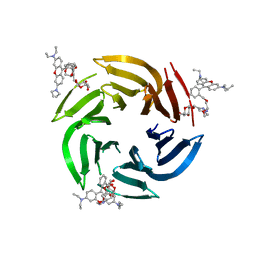 | | Crystal structure of RSL mutant in complex with Ligand | | Descriptor: | 2-[2-[4-[[(2R,3S,4R,5S,6S)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-1,2,3-triazol-1-yl]ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, Fucose-binding lectin protein,Fucose-binding lectin protein,Fucose-binding lectin protein | | Authors: | Li, L, Chen, G.S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of RSL mutant-R17A/R108A/R199A in complex with R3F
To Be Published
|
|
4DYA
 
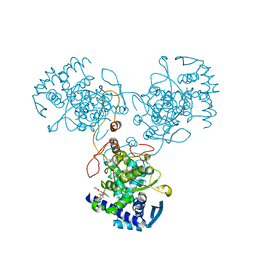 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-885986 Ligand Bound | | Descriptor: | N-[4-chloranyl-5-[4-[[3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl]piperazin-1-yl]-2-nitro-phenyl]furan-2-carboxamide, Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | To be determined
To be Published
|
|
4DYP
 
 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-831780 Ligand Bound | | Descriptor: | Nucleocapsid protein, [4-(5-bromanyl-3-methyl-pyridin-2-yl)piperazin-1-yl]-[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | To be determined
To be Published
|
|
4YDN
 
 | | High resolution crystal structure of human transthyretin bound to ligand and conjugates of 4-(5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl)phenyl fluorosulfate | | Descriptor: | 2,6-dichloro-4-[5-(4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl]phenol, 4-[5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl]phenyl sulfurofluoridate, Transthyretin | | Authors: | Connelly, S, Bradbury, N.C. | | Deposit date: | 2015-02-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Fluorogenic Aryl Fluorosulfate for Intraorganellar Transthyretin Imaging in Living Cells and in Caenorhabditis elegans.
J.Am.Chem.Soc., 137, 2015
|
|
5HPF
 
 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain from Acinetobacter | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
7JQ5
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
4DHL
 
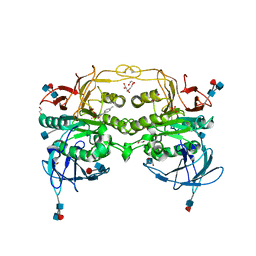 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment MO07123 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Clayton, D.J, Hussein, W.M, Schenk, G, McGeary, R, Guddat, L.W. | | Deposit date: | 2012-01-29 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
7JQ3
 
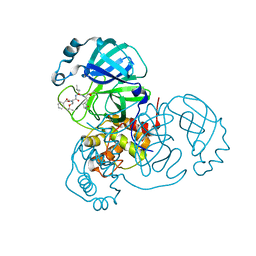 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI6 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
2Q8M
 
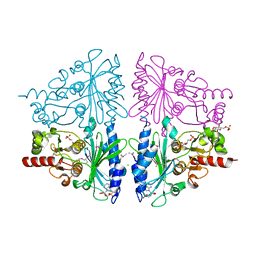 | | T-like Fructose-1,6-bisphosphatase from Escherichia coli with AMP, Glucose 6-phosphate, and Fructose 1,6-bisphosphate bound | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Hines, J.K, Kruesel, C.E, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Inhibited Fructose-1,6-bisphosphatase from Escherichia coli: DISTINCT ALLOSTERIC INHIBITION SITES FOR AMP AND GLUCOSE 6-PHOSPHATE AND THE CHARACTERIZATION OF A GLUCONEOGENIC SWITCH.
J.Biol.Chem., 282, 2007
|
|
6VVV
 
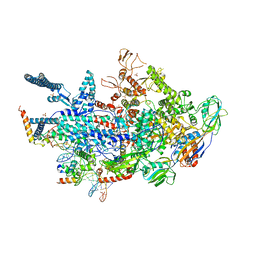 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2MCC
 
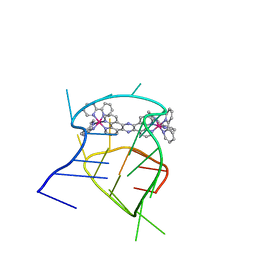 | | Structural studies on dinuclear ruthenium(II) complexes that bind diastereoselectively to an anti-parallel folded human telomere sequence | | Descriptor: | human_telomere_quadruplex, tetrakis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)(mu-tetrapyrido[3,2-a:2',3'-c:3'',2''-h:2''',3'''-j]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~13~,N~14~)diruthenium(4+) | | Authors: | Williamson, M.P, Wilson, T, Thomas, J.A, Felix, V, Costa, P.J. | | Deposit date: | 2013-08-18 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence.
J.Med.Chem., 56, 2013
|
|
7NEI
 
 | |
2MYC
 
 | |
2MYA
 
 | |
4TOP
 
 | | Glycine max glutathione transferase | | Descriptor: | 2,4-D inducible glutathione S-transferase, GLUTATHIONE | | Authors: | Axarli, I, Dhavala, P, Papageorgiou, A.C. | | Deposit date: | 2014-06-06 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Comparative analysis of the structural and functional features of two homologous tau class glutathione transferases from Glycine max
To Be Published
|
|
