8H2H
 
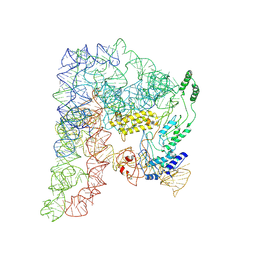 | | Cryo-EM structure of a Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | Group II intron-encoded protein LtrA, LtrB, RNA (5'-R(P*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*C)-3') | | Authors: | Liu, N, Dong, X.L, Qu, G.S, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Functionalized graphene grids with various charges for single-particle cryo-EM.
Nat Commun, 13, 2022
|
|
8B9L
 
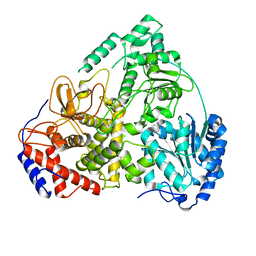 | | Cryo-EM structure of MLE | | Descriptor: | Dosage compensation regulator | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
8B9J
 
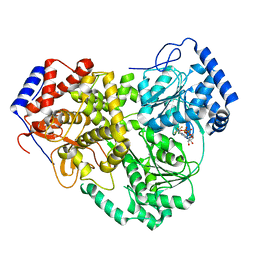 | | Cryo-EM structure of MLE in complex with ADP:AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dosage compensation regulator, MAGNESIUM ION, ... | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
8B9K
 
 | |
8EPC
 
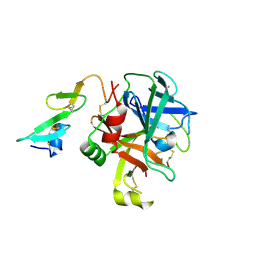 | |
8EPK
 
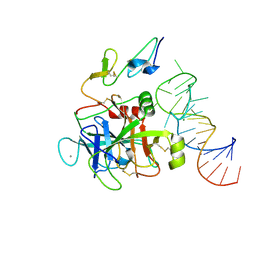 | |
8EPH
 
 | | Crystal structure of human coagulation factor IXa (S195A), apo-form, DES-GLA | | Descriptor: | CALCIUM ION, Coagulation factor IXa heavy chain, Coagulation factor IXa light chain, ... | | Authors: | Kolyadko, V.N, Krishnaswamy, S. | | Deposit date: | 2022-10-05 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An RNA aptamer exploits exosite-dependent allostery to achieve specific inhibition of coagulation factor IXa.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8B9F
 
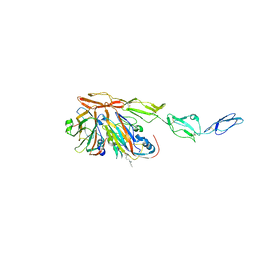 | | Structure of Echovirus 11 complexed with DAF (CD55) calculated from symmetry expansion | | Descriptor: | Complement decay-accelerating factor, Genome polyprotein, SPHINGOSINE | | Authors: | Stuart, D.I, Ren, J, Qin, L, Zhou, D. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
8B8S
 
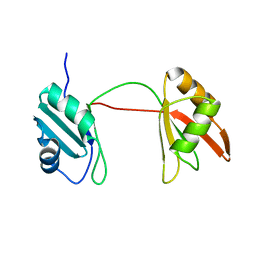 | | Solution structure of tandem RRM1 and RRM2 domains of yeast NPL3 | | Descriptor: | Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 | | Authors: | Kachariya, N, Sattler, M, Keil, P, Strasser, K. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res., 51, 2023
|
|
8B8R
 
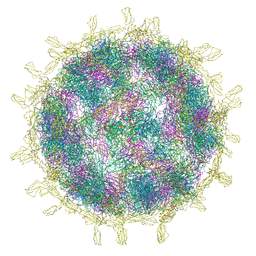 | | Complex of Echovirus 11 with its attaching receptor decay-accelerating factor (CD55) | | Descriptor: | DECAY ACCELERATING FACTOR (CD55), SPHINGOSINE, VP1, ... | | Authors: | Stuart, D.I, Ren, J, Zhou, D, Qin, L. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-07 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
8EP1
 
 | |
8EOW
 
 | |
8EP0
 
 | |
8EOP
 
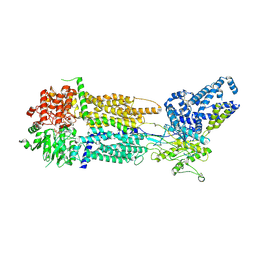 | | Cryo-EM Structure of Nanodisc reconstituted human ABCA7 EQ mutant in ATP bound closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EOJ
 
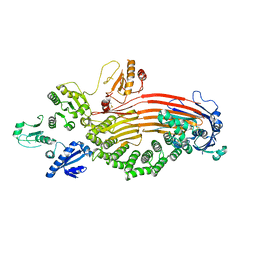 | | Microsomal triglyceride transfer protein | | Descriptor: | Microsomal triglyceride transfer protein large subunit, Protein disulfide-isomerase | | Authors: | Zhang, Z. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EOG
 
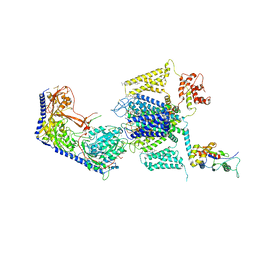 | | Structure of the human L-type voltage-gated calcium channel Cav1.2 complexed with L-leucine | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl dodecanoate, (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl heptadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Mondal, A, Abderemane-Ali, F, Minor, D.L. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | EMC chaperone-Ca V structure reveals an ion channel assembly intermediate.
Nature, 619, 2023
|
|
8H1O
 
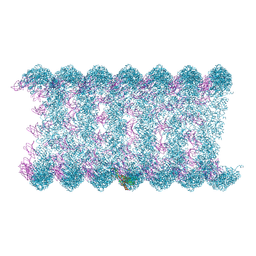 | | Cryo-EM structure of KpFtsZ-monobody double helical tube | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Mb(Ec/KpFtsZ_S1) | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8EOH
 
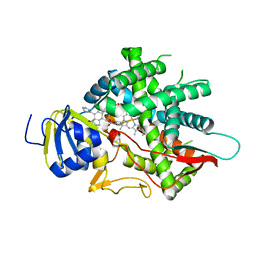 | | crystal structure of human Cytochrome P450 8B1 in complex with a C12-Pyridine Containing Steroid | | Descriptor: | 12-(pyridin-3-yl)-8alpha,10alpha,13alpha,14beta-androsta-4,11-diene-3,17-dione, 7-alpha-hydroxycholest-4-en-3-one 12-alpha-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, J, Scott, E.E. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Pyridine-containing substrate analogs are restricted from accessing the human cytochrome P450 8B1 active site by tryptophan 281.
J.Biol.Chem., 299, 2023
|
|
8EOK
 
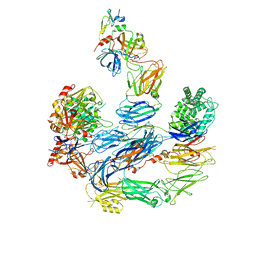 | |
8H0R
 
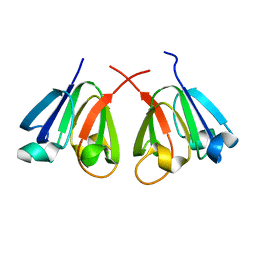 | |
8H0P
 
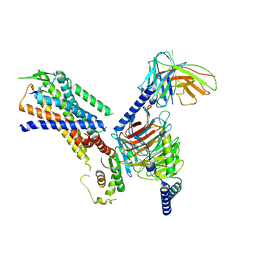 | | Structure of the NMB30-NMBR and Gq complex | | Descriptor: | G-alpha q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Xu, Y, Liu, H, Cai, H, Xu, H.E, Yin, W. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular recognition of itch-associated neuropeptides by bombesin receptors
Cell Res., 33, 2023
|
|
8H0Q
 
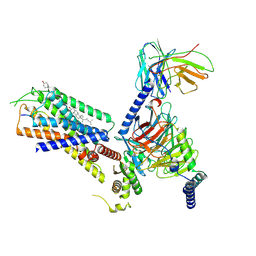 | | Structure of the GRP14-27-GRPR-Gq complex | | Descriptor: | CHOLESTEROL, G-alpha q, GRP, ... | | Authors: | Li, C, Xu, Y, Liu, H, Cai, H, Xu, H.E, Yin, W. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of itch-associated neuropeptides by bombesin receptors
Cell Res., 33, 2023
|
|
8ENU
 
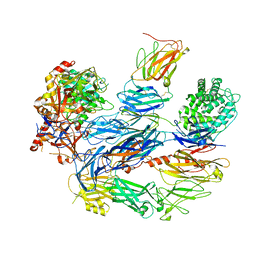 | | Structure of the C3bB proconvertase in complex with lufaxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 beta chain, Complement C3b alpha' chain, ... | | Authors: | Andersen, J.F, Lei, H. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bispecific inhibitor of complement and coagulation blocks activation in complementopathy models via a novel mechanism.
Blood, 141, 2023
|
|
8ENJ
 
 | |
8B7Q
 
 | | Cryo-EM structure for the mouse LEPR-CRH2:Leptin:LEPR-Ig complex following symmetry expansion in combination with local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-09-30 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
