1ZTQ
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with WAY-033 | | Descriptor: | CALCIUM ION, Collagenase 3, N-({4'-[(1-BENZOFURAN-2-YLCARBONYL)AMINO]-1,1'-BIPHENYL-4-YL}SULFONYL)-L-VALINE, ... | | Authors: | Wu, J, Rush III, T.S, Hotchandani, R, Du, X, Geck, M, Collins, E, Xu, Z.B, Skotnicki, J, Levin, J.I, Lovering, F. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of potent and selective MMP-13 inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1ZTR
 
 | | Solution structure of Engrailed homeodomain L16A mutant | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L, Markson, J.S, Mayor, U, Freund, S.M.V, Fersht, A.R. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a protein denatured state and folding intermediate.
Nature, 437, 2005
|
|
1ZTS
 
 | | Solution Structure of Bacillus Subtilis Protein YQBG: Northeast Structural Genomics Consortium Target SR215 | | Descriptor: | Hypothetical protein yqbG | | Authors: | Liu, G, Shen, Y, Xiao, R, Acton, T, Ma, L, Joachimiak, A, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-27 | | Release date: | 2005-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein yqbG from Bacillus subtilis reveals a novel alpha-helical protein fold.
Proteins, 62, 2006
|
|
1ZTT
 
 | | Netropsin bound to d(CTTAATTCGAATTAAG) in complex with MMLV RT catalytic fragment | | Descriptor: | 5'-D(*CP*TP*TP*AP*AP*TP*TP*C)-3', 5'-D(P*GP*AP*AP*TP*TP*AP*AP*G)-3', NETROPSIN, ... | | Authors: | Goodwin, K.D, Long, E.C, Georgiadis, M.M. | | Deposit date: | 2005-05-27 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A host-guest approach for determining drug-DNA interactions: an example using netropsin.
Nucleic Acids Res., 33, 2005
|
|
1ZTU
 
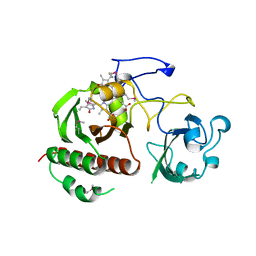 | | Structure of the chromophore binding domain of bacterial phytochrome | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Wagner, J.R, Brunzelle, J.S, Forest, K.T, Vierstra, R.D. | | Deposit date: | 2005-05-27 | | Release date: | 2005-11-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome.
Nature, 438, 2005
|
|
1ZTV
 
 | |
1ZTW
 
 | |
1ZTX
 
 | | West Nile Virus Envelope Protein DIII in complex with neutralizing E16 antibody Fab | | Descriptor: | Envelope protein, Heavy Chain of E16 Antibody, Light Chain of E16 Antibody | | Authors: | Nybakken, G.E, Oliphant, T, Diamond, M.S, Fremont, D.H. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of West Nile virus neutralization by a therapeutic antibody.
Nature, 437, 2005
|
|
1ZTY
 
 | |
1ZTZ
 
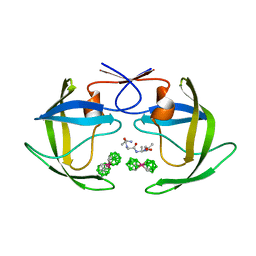 | | Crystal structure of HIV protease- metallacarborane complex | | Descriptor: | COBALT BIS(1,2-DICARBOLLIDE), PROTEASE RETROPEPSIN, autoproteolytic tetrapeptide | | Authors: | Cigler, P, Kozisek, M, Rezacova, P, Brynda, J, Otwinowski, Z, Sedlacek, J, Bodem, J, Kraeusslich, H.-G, Kral, V, Konvalinka, J. | | Deposit date: | 2005-05-28 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | From nonpeptide toward noncarbon protease inhibitors: Metallacarboranes as specific and potent inhibitors of HIV protease
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1ZU0
 
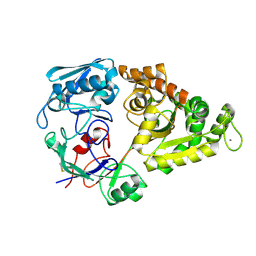 | |
1ZU1
 
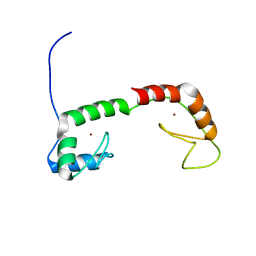 | | Solution Structure of the N-terminal Zinc Fingers of the Xenopus laevis double stranded RNA binding protein ZFa | | Descriptor: | RNA binding protein ZFa, ZINC ION | | Authors: | Moller, H.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-05-29 | | Release date: | 2005-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc fingers of the Xenopus laevis double-stranded RNA-binding protein ZFa
J.Mol.Biol., 351, 2005
|
|
1ZU2
 
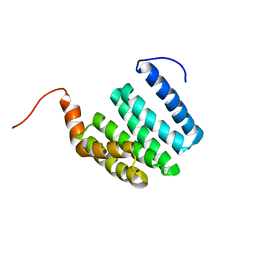 | |
1ZU3
 
 | | Crystal Structure Of Mutant K8A Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-05-30 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1ZU4
 
 | |
1ZU5
 
 | |
1ZU8
 
 | | Crystal structure of the goat signalling protein with a bound trisaccharide reveals that Trp78 reduces the carbohydrate binding site to half | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3 like protein 1, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ethayathulla, A.S, Kumar, J, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2005-05-30 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the goat signalling protein with a bound trisaccharide reveals that Trp78 reduces the carbohydrate binding site to half
To be Published
|
|
1ZUA
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Tolrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT | | Authors: | Gallego, O, Ruiz, F.X, Ardevol, A, Dominguez, M, Alvarez, R, de Lera, A.R, Rovira, C, Farres, J, Fita, I, Pares, X. | | Deposit date: | 2005-05-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1ZUB
 
 | | Solution Structure of the RIM1alpha PDZ Domain in Complex with an ELKS1b C-terminal Peptide | | Descriptor: | ELKS1b, Regulating synaptic membrane exocytosis protein 1 | | Authors: | Lu, J, Li, H, Wang, Y, Sudhof, T.C, Rizo, J. | | Deposit date: | 2005-05-30 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RIM1alpha PDZ Domain in Complex with an ELKS1b C-terminal Peptide
J.Mol.Biol., 352, 2005
|
|
1ZUC
 
 | | Progesterone receptor ligand binding domain in complex with the nonsteroidal agonist tanaproget | | Descriptor: | 5-(4,4-DIMETHYL-2-THIOXO-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-6-YL)-1-METHYL-1H-PYRROLE-2-CARBONITRILE, Progesterone receptor, SULFATE ION | | Authors: | Zhang, Z, Olland, A.M, Zhu, Y, Cohen, J, Berrodin, T, Chippari, S, Appavu, C, Li, S, Wilhem, J, Chopra, R, Fensome, A, Zhang, P, Wrobel, J, Unwalla, R.J, Lyttle, C.R, Winneker, R.C. | | Deposit date: | 2005-05-30 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and pharmacological properties of a potent and selective novel nonsteroidal progesterone receptor agonist tanaproget
J.Biol.Chem., 280, 2005
|
|
1ZUD
 
 | | Structure of ThiS-ThiF protein complex | | Descriptor: | Adenylyltransferase thiF, CALCIUM ION, SODIUM ION, ... | | Authors: | Ealick, S.E, Lehmann, C. | | Deposit date: | 2005-05-30 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Escherichia coli ThiS-ThiF Complex, a Key Component of the Sulfur Transfer System in Thiamin Biosynthesis.
Biochemistry, 45, 2006
|
|
1ZUE
 
 | | Revised Solution Structure of DLP-2 | | Descriptor: | Defensin-like peptide 2/4 | | Authors: | Torres, A.M, Tsampazi, C, Geraghty, D.P, Bansal, P.S, Alewood, P.F, Kuchel, P.W. | | Deposit date: | 2005-05-30 | | Release date: | 2005-08-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | D-amino acid residue in a defensin-like peptide from platypus venom: effect on structure and chromatographic properties.
Biochem.J., 391, 2005
|
|
1ZUF
 
 | | Solution Structure of DLP-4 | | Descriptor: | Defensin-like peptide 2/4 | | Authors: | Torres, A.M, Tsampazi, C, Geraghty, D.P, Bansal, P.S, Alewood, P.F, Kuchel, P.W. | | Deposit date: | 2005-05-31 | | Release date: | 2005-08-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | D-amino acid residue in a defensin-like peptide from platypus venom: effect on structure and chromatographic properties.
Biochem.J., 391, 2005
|
|
1ZUG
 
 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
1ZUH
 
 | |
