1U9M
 
 | | Crystal structure of F58W mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1CNM
 
 | | ENHANCEMENT OF CATALYTIC EFFICIENCY OF PROTEINASE K THROUGH EXPOSURE TO ANHYDROUS ORGANIC SOLVENT AT 70 DEGREES CELSIUS | | Descriptor: | ACETONITRILE, CALCIUM ION, PROTEIN (PROTEINASE K) | | Authors: | Gupta, M.N, Tyagi, R, Sharma, S, Karthikeyan, S, Singh, T.P. | | Deposit date: | 1999-05-20 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enhancement of catalytic efficiency of enzymes through exposure to anhydrous organic solvent at 70 degrees C. Three-dimensional structure of a treated serine proteinase at 2.2 A resolution.
Proteins, 39, 2000
|
|
3F4A
 
 | | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family | | Descriptor: | AMMONIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Osipiuk, J, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family
To be Published
|
|
1U9U
 
 | | Crystal structure of F58Y mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3FZZ
 
 | | Structure of GrC | | Descriptor: | Granzyme C, SULFATE ION | | Authors: | Buckle, A.M, Kaiserman, D, Whisstock, J.C. | | Deposit date: | 2009-01-27 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of granzyme C reveals an unusual mechanism of protease autoinhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3F4F
 
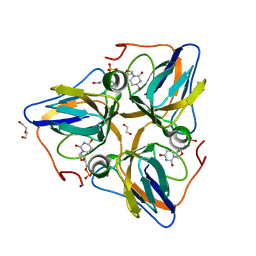 | | Crystal structure of dUT1p, a dUTPase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3GIS
 
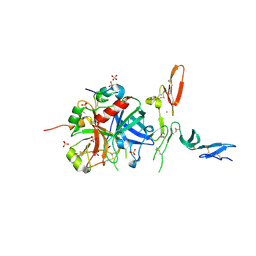 | |
2KNP
 
 | |
2O8U
 
 | | Crystal Structure and Binding Epitopes of Urokinase-type Plasminogen Activator (C122A/N145Q/S195A) in complex with Inhibitors | | Descriptor: | BENZAMIDINE, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Zhao, G, Yuan, C, Jiang, L, Huang, Z, Huang, M. | | Deposit date: | 2006-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure and Binding Epitopes of Urokinase-type Plasminogen Activator (C122A/N145Q/S195A) in complex with Inhibitors
To be Published
|
|
2O8W
 
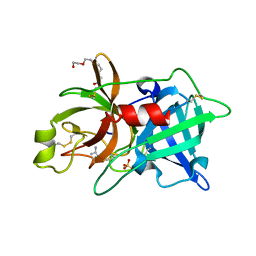 | | Crystal Structure and Binding Epitopes of Urokinase-type Plasminogen Activator (C122A/N145Q/S195A) in complex with Inhibitors | | Descriptor: | 1-phenylguanidine, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Zhao, G, Yuan, C, Jiang, L, Huang, Z, Huang, M. | | Deposit date: | 2006-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure and Binding Epitopes of Urokinase-type Plasminogen Activator (C122A/N145Q/S195A) in complex with Inhibitors
To be Published
|
|
6RUR
 
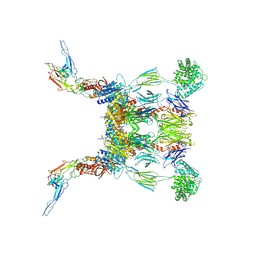 | | Structure of the SCIN stabilized C3bBb convertase bound to properdin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, ... | | Authors: | Pedersen, D.V, Gadeberg, T.A.F, Andersen, G.R. | | Deposit date: | 2019-05-29 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural Basis for Properdin Oligomerization and Convertase Stimulation in the Human Complement System.
Front Immunol, 10, 2019
|
|
6RUV
 
 | |
6TS4
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
1O6G
 
 | |
1O6F
 
 | |
7WQZ
 
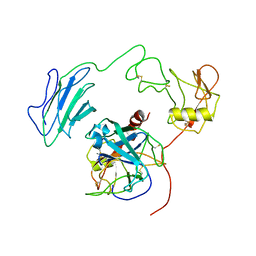 | | Structure of Active-mutEP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Enteropeptidase catalytic light chain, Enteropeptidase non-catalytic heavy chain | | Authors: | Yang, X.L, Ding, Z.Y, Huang, H.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase.
Nat Commun, 13, 2022
|
|
7XKC
 
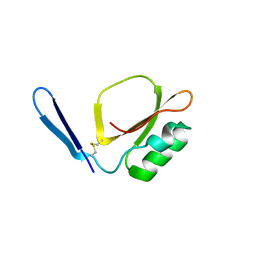 | | Crystal structure of Daucus Carrot hypoglycemic peptide (DCHP) | | Descriptor: | DCHP, SULFATE ION | | Authors: | Guo, T, Ren, J.Q, Wang, L, Shi, Y.W, Feng, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Characterization of a thermostable, protease-tolerant inhibitor of alpha-glycosidase from carrot: A potential oral additive for treatment of diabetes.
Int.J.Biol.Macromol., 209, 2022
|
|
8DCN
 
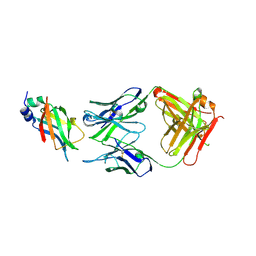 | |
8DCM
 
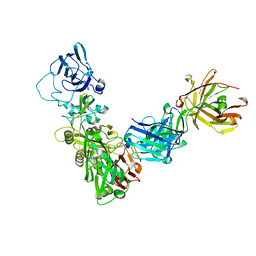 | |
3KCG
 
 | | Crystal structure of the antithrombin-factor IXa-pentasaccharide complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, Antithrombin-III, ... | | Authors: | Huntington, J.A, Johnson, D.J.D. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of factor IXa recognition by heparin-activated antithrombin revealed by a 1.7-A structure of the ternary complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8SRZ
 
 | |
3LD6
 
 | | Crystal structure of human lanosterol 14alpha-demethylase (CYP51) in complex with ketoconazole | | Descriptor: | 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Lanosterol 14-alpha demethylase, ... | | Authors: | Strushkevich, N, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of human CYP51 inhibition by antifungal azoles.
J.Mol.Biol., 397, 2010
|
|
3LU9
 
 | | Crystal structure of human thrombin mutant S195A in complex with the extracellular fragment of human PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Proteinase-activated receptor 1, ... | | Authors: | Gandhi, P.S, Chen, Z, Di Cera, E. | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of thrombin bound to the uncleaved extracellular fragment of PAR1.
J.Biol.Chem., 285, 2010
|
|
3JUV
 
 | | Crystal structure of human lanosterol 14alpha-demethylase (CYP51) | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Strushkevich, N, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Usanov, S.A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis of human CYP51 inhibition by antifungal azoles.
J. Mol. Biol., 397, 2010
|
|
3JUS
 
 | | Crystal structure of human lanosterol 14alpha-demethylase (CYP51) in complex with econazole | | Descriptor: | 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, 1-[(2S)-2-[(4-CHLOROBENZYL)OXY]-2-(2,4-DICHLOROPHENYL)ETHYL]-1H-IMIDAZOLE, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Strushkevich, N, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Usanov, S.A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human CYP51 inhibition by antifungal azoles.
J. Mol. Biol., 397, 2010
|
|
