4FHC
 
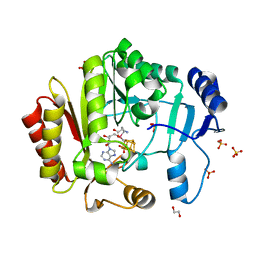 | | Spore photoproduct lyase | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
6UMD
 
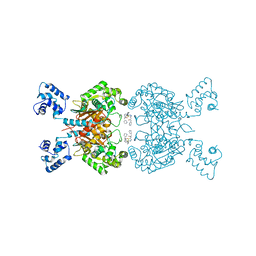 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-(pyridin-3-yl)-N-(5-{4-[(5-{[(pyridin-3-yl)acetyl]amino}-1,3,4-thiadiazol-2-yl)amino]piperidin-1-yl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
5YCP
 
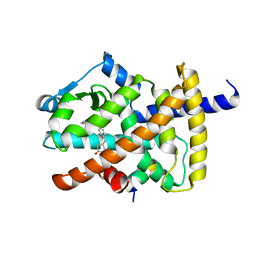 | | Human PPARgamma ligand binding domain complexed with Rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Nuclear receptor coactivator 1, ... | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPAR gamma.
Sci Rep, 8, 2018
|
|
7F6R
 
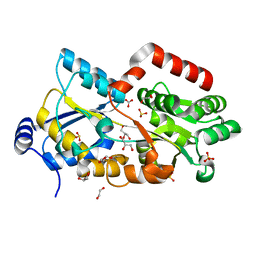 | |
7BN5
 
 | |
4KA4
 
 | |
7F6N
 
 | |
7BI2
 
 | | PI3KC2aDeltaN and DeltaC-C2 | | Descriptor: | 1,2-ETHANEDIOL, 9-(6-aminopyridin-3-yl)-1-[3-(trifluoromethyl)phenyl]benzo[h][1,6]naphthyridin-2(1H)-one, IODIDE ION, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
9I6J
 
 | | Hen egg-white lysozyme structure embedded in LCP medium at 95% relative humidity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Zabelskii, D, Round, E, Han, H, von Stetten, D, Melo, D, de Wijn, R, Round, A. | | Deposit date: | 2025-01-30 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Viscoelastic characterization of the lipid cubic phase provides insights into high-viscosity extrusion injection for XFEL experiments.
To Be Published
|
|
5HJ9
 
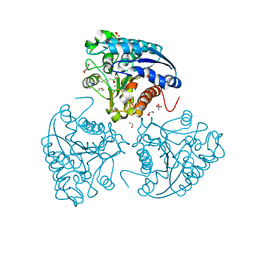 | | Crystal structure of Leishmania mexicana arginase in complex with inhibitor ABHPE | | Descriptor: | 1,2-ETHANEDIOL, Arginase, GLYCEROL, ... | | Authors: | Hai, Y, Christianson, D.W. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of Leishmania mexicana arginase complexed with alpha , alpha-disubstituted boronic amino-acid inhibitors.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4BWA
 
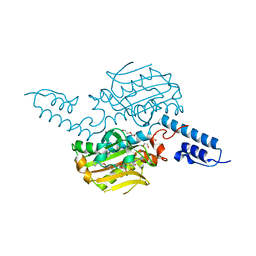 | | PylRS Y306G, Y384F, I405R mutant in complex with adenylated norbornene | | Descriptor: | 1,2-ETHANEDIOL, Adenylated Norbornene, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schneider, S, Vrabel, M, Gattner, M.J, Fluegel, V, Lopez-Carillo, V, Carell, T. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Insights Into Incorporation of Norbornene Amino Acids for Click Modification of Proteins
Chem.Bio.Chem., 14, 2013
|
|
9I6M
 
 | | Hen egg-white lysozyme structure at 65% relative humidity | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Zabelskii, D, Round, E, Han, H, von Stetten, D, Melo, D, de Wijn, R, Bean, R, Round, A. | | Deposit date: | 2025-01-30 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Viscoelastic characterization of the lipid cubic phase provides insights into high-viscosity extrusion injection for XFEL experiments.
To Be Published
|
|
8XMG
 
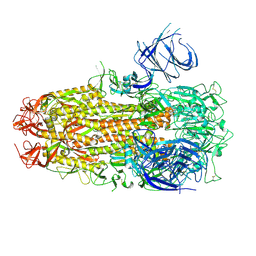 | | Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P), RBD-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XMT
 
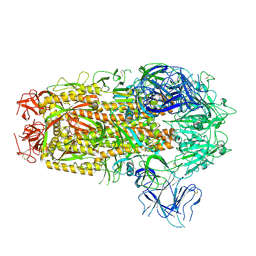 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P), RBD-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-28 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
1LAQ
 
 | | Solution Structure of the B-DNA Duplex CGCGGTXTCCGCG (X=PdG) Containing the 1,N2-propanodeoxyguanosine Adduct with the Deoxyribose at C20 Opposite PdG in the C2' Endo Conformation. | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*(DNR)P*AP*CP*CP*GP*CP*G)-3', 5'-D(*CP*GP*CP*GP*GP*TP*(P)P*TP*CP*CP*GP*CP*G)-3' | | Authors: | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | Deposit date: | 2002-03-29 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an oligodeoxynucleotide containing a 1,N(2)-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2.
Chem.Res.Toxicol., 15, 2002
|
|
5KW2
 
 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | Descriptor: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | Authors: | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | Deposit date: | 2016-07-15 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
6KX8
 
 | | Crystal structure of mouse Cryptochrome 2 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-2 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Isoform-selective regulation of mammalian cryptochromes.
Nat.Chem.Biol., 16, 2020
|
|
4OBQ
 
 | | MAP4K4 in complex with inhibitor (compound 31), N-[3-(4-AMINOQUINAZOLIN-6-YL)-5-FLUOROPHENYL]-2-(PYRROLIDIN-1-YL)ACETAMIDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Mitogen-activated protein kinase kinase kinase kinase 4, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-01-07 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Selective 4-Amino-pyridopyrimidine Inhibitors of MAP4K4 Using Fragment-Based Lead Identification and Optimization.
J.Med.Chem., 57, 2014
|
|
5L30
 
 | | Factor VIIa in complex with the inhibitor (2R,15R)-2-[(1-aminoisoquinolin-6-yl)amino]-4,15,17-trimethyl-7-[1-(1H-tetrazol-5-yl)cyclopropyl]-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaene-3,12-dione | | Descriptor: | (2R,15R)-2-[(1-aminoisoquinolin-6-yl)amino]-4,15,17-trimethyl-7-[1-(1H-tetrazol-5-yl)cyclopropyl]-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaene-3,12-dione, CALCIUM ION, Coagulation factor VII (Heavy Chain), ... | | Authors: | Wei, A. | | Deposit date: | 2016-08-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Synthesis and P1' SAR exploration of potent macrocyclic tissue factor-factor VIIa inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5UIS
 
 | | Crystal structure of IRAK4 in complex with compound 12 | | Descriptor: | 4-{[(3R)-piperidin-3-yl]oxy}-6-[(propan-2-yl)oxy]quinoline-7-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
6UO6
 
 | |
7YHC
 
 | | Crystal structure of VIM-2 MBL in complex with 3-(4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl)phthalic acid | | Descriptor: | 3-[4-(3-aminophenyl)-1,2,3-triazol-1-yl]phthalic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Metal binding pharmacophore click-derived discovery of new broad-spectrum metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 257, 2023
|
|
9HWV
 
 | | Crystal structure of the Keap1 Kelch domain in complex with the small molecule UCAB#1010 at 1.50 Angstrom resolution | | Descriptor: | (3~{S})-3-(1,3-benzodioxol-5-yl)-3-[7-[2-[[(1~{R})-cyclohex-2-en-1-yl]amino]-2-oxidanylidene-ethoxy]naphthalen-2-yl]propanoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2025-01-06 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment-Based Drug Discovery of Novel High-affinity, Selective, and Anti-inflammatory Inhibitors of the Keap1-Nrf2 Protein-Protein Interaction.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
6L4V
 
 | |
5UIQ
 
 | | Crystal structure of IRAK4 in complex with compound 9 | | Descriptor: | 2-[(propan-2-yl)oxy]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
