6IHE
 
 | |
6PDJ
 
 | | Tyrosine-protein kinase LCK bound to Compound 11 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, N-{4-[(6-methoxypyrazolo[1,5-a]pyridine-3-carbonyl)amino]-3-methylphenyl}-1-methyl-1H-indazole-3-carboxamide, NICKEL (II) ION, ... | | Authors: | Critton, D.A. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Pyridazinone and Pyrazolo[1,5-a]pyridine Inhibitors of C-Terminal Src Kinase.
Acs Med.Chem.Lett., 10, 2019
|
|
9HB0
 
 | | Crystal structure of Plasmodium falciparum Plasmepsin X in complex with the hydroxyethylamine drug 7k. | | Descriptor: | (4S)-4-[(1R)-2-[2-(3-methoxyphenyl)propan-2-ylamino]-1-oxidanyl-ethyl]-16-propyl-3,16-diazatricyclo[16.3.1.1^{6,10}]tricosa-1(21),6,8,10(23),18(22),19-hexaene-2,17-dione, Plasmepsin X, SULFATE ION | | Authors: | Withers-Martinez, C, George, R, Ogrodowicz, R, Kunzelmann, S, Purkiss, A, Kjaer, S, Walker, P, Kovada, V, Jirgensons, A, Blackman, M.J. | | Deposit date: | 2024-11-05 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Plasticity of Plasmodium falciparum Plasmepsin X to Accommodate Binding of Potent Macrocyclic Hydroxyethylamine Inhibitors.
J.Mol.Biol., 437, 2025
|
|
6EQB
 
 | | HLA class I histocompatibility antigen | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALA-ALA-GLY-ILE-GLY-ILE-LEU-THR-VAL, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | TCR-induced alteration of primary MHC peptide anchor residue.
Eur.J.Immunol., 49, 2019
|
|
6TCU
 
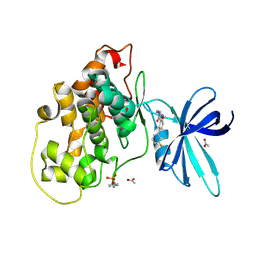 | | Glycogen synthase kinase-3 beta (GSK3b) in complex with ligand 1 | | Descriptor: | 5-[2,3-bis(fluoranyl)phenyl]-~{N}-[[1-(2-methoxyethyl)piperidin-4-yl]methyl]-1~{H}-indazole-3-carboxamide, ACETATE ION, Glycogen synthase kinase-3 beta | | Authors: | Lammens, A, Krapp, S, Buonfiglio, R, Ombrato, R. | | Deposit date: | 2019-11-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of Indazole-Based GSK-3 Inhibitors with Mitigated hERG Issue andIn VivoActivity in a Mood Disorder Model.
Acs Med.Chem.Lett., 11, 2020
|
|
7CWG
 
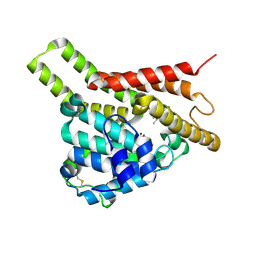 | | Crystal structure of PDE8A catalytic domain in complex with 3a | | Descriptor: | 9-[(1~{S})-1-[4-[2,2-bis(fluoranyl)ethoxy]pyridin-2-yl]ethyl]-2-chloranyl-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Huang, Y, Wu, X.-N, Zhou, Q, Wu, Y, Luo, H.-B. | | Deposit date: | 2020-08-28 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rational Design of 2-Chloroadenine Derivatives as Highly Selective Phosphodiesterase 8A Inhibitors.
J.Med.Chem., 63, 2020
|
|
5ABG
 
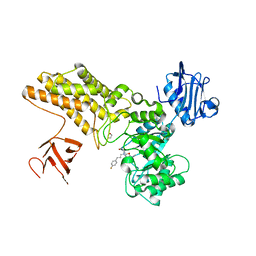 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-1-[3-(4-fluorophenyl)propyl]-5-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7VH6
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the active state (pH 6.0, BeF3-, conformation 1, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BERYLLIUM TRIFLUORIDE ION, Plasma membrane ATPase 1 | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
3O84
 
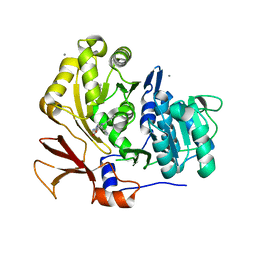 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
8JHR
 
 | | Cryo-EM structure of human S1P transporter SPNS2 bound with an inhibitor 16d | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Pang, B, Yu, L.Y, Ren, R.B. | | Deposit date: | 2023-05-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Molecular basis of Spns2-facilitated sphingosine-1-phosphate transport.
Cell Res., 34, 2024
|
|
9INF
 
 | | Cryo-EM structure of human XPR1 in closed state in the presence of KIDINS220-1-432 and 10 mM KH2PO4 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ARACHIDONIC ACID, CHOLESTEROL, ... | | Authors: | Yin, Y, Zuo, P, Liang, L. | | Deposit date: | 2024-07-06 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
6BN3
 
 | | CTX-M-151 class A extended-spectrum beta-lactamase apo crystal structure at 1.3 Angstrom resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase | | Authors: | Power, P, Ghiglione, B, Rodriguez, M.M, Gutkind, G, Ishii, Y, Bonomo, R.A, Klinke, S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.278 Å) | | Cite: | Structural and Biochemical Characterization of the Novel CTX-M-151 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam.
Antimicrob.Agents Chemother., 65, 2021
|
|
9INH
 
 | | Cryo-EM structure of human XPR1 in complex with InsP6 in outward-facing state (SPX visible)-in the presence of KIDINS220-1-432 and 10 mM KH2PO4 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ARACHIDONIC ACID, CHOLESTEROL, ... | | Authors: | Zuo, P, Liang, L, Yin, Y. | | Deposit date: | 2024-07-06 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
9IUC
 
 | | Cryo-EM structure of human XPR1 in complex with InsP6 in closed state - in the presence of KIDINS220-1-432 without substrate KH2PO4 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ARACHIDONIC ACID, CHOLESTEROL, ... | | Authors: | Zuo, P, Liang, L, Yin, Y. | | Deposit date: | 2024-07-20 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Synergistic activation of the human phosphate exporter XPR1 by KIDINS220 and inositol pyrophosphate.
Nat Commun, 16, 2025
|
|
6EDM
 
 | | Structure of apo-CDD-1 beta-lactamase | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-14 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of CDD-1, the intrinsic class D beta-lactamase from the pathogenic Gram-positive bacterium Clostridioides difficile, and its complex with cefotaxime.
J.Struct.Biol., 208, 2019
|
|
1PLW
 
 | |
6ITM
 
 | | Crystal structure of FXR in complex with agonist XJ034 | | Descriptor: | 1-adamantyl-[4-(5-chloranyl-2-methyl-phenyl)piperazin-1-yl]methanone, Bile acid receptor, HD3 Peptide from Nuclear receptor coactivator 1 | | Authors: | Zhang, H, Wang, Z. | | Deposit date: | 2018-11-23 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pose Filter-Based Ensemble Learning Enables Discovery of Orally Active, Nonsteroidal Farnesoid X Receptor Agonists.
J.Chem.Inf.Model., 60, 2020
|
|
6RJP
 
 | | Bfl-1 in complex with alpha helical peptide | | Descriptor: | Bcl-2-like protein 11, Bcl-2-related protein A1 | | Authors: | Baggio, C, Gambini, L, Udompholkul, P, Salem, A.F, Hakansson, M, Jossart, J, Perry, J, Pellecchia, M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | N-locking stabilization of covalent helical peptides: Application to Bfl-1 antagonists.
Chem.Biol.Drug Des., 95, 2020
|
|
3KRO
 
 | | Mint heterotetrameric geranyl pyrophosphate synthase in complex with magnesium, IPP, and DMASPP (II) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Chang, T.-H, Hsieh, F.-L, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2009-11-19 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation
Plant Cell, 22, 2010
|
|
3KQQ
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 2-Hydroxynicotinic acid | | Descriptor: | 2-oxo-1,2-dihydropyridine-3-carboxylic acid, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
6RGA
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with an inhibitor | | Descriptor: | (2~{R},3~{S},4~{R},5~{R})-2-(aminomethyl)-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy]prop-1-ynyl]-6-azanyl-purin-9-yl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | From Substrate to Fragments to Inhibitor ActiveIn VivoagainstStaphylococcus aureus.
Acs Infect Dis., 6, 2020
|
|
6V7E
 
 | | Human Arginase1 Complexed with Bicyclic Inhibitor Compound 12 | | Descriptor: | 3-[(5~{S},7~{S},8~{S})-8-azanyl-8-carboxy-1-azaspiro[4.4]nonan-7-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2019-12-08 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery and Optimization of Rationally Designed Bicyclic Inhibitors of Human Arginase to Enhance Cancer Immunotherapy.
Acs Med.Chem.Lett., 11, 2020
|
|
3MNQ
 
 | | Crystal structure of myosin-2 motor domain in complex with ADP-metavanadate and resveratrol | | Descriptor: | 1,2-ETHANEDIOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Schneider, J, Taft, M, Backhaus, A, Baruch, P, Fedorov, R, Manstein, D.J. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of resveratrol regulation of myosin activity.
To be Published
|
|
6FVF
 
 | | The Structure of CK2alpha with CCh503 bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, [1-[2-(phenylsulfonylamino)ethyl]piperidin-4-yl]methyl 5-fluoranyl-2-methoxy-1~{H}-indole-3-carboxylate | | Authors: | Brear, P, Prudent, R, Laudet, B, Filhol, O, Cochet, C, Sautel, C, Moucadel, V, Bestgen, B, Engel, M, Ettaoussi, M, Lomberget, T, Le Borgne, M, Kufareva, I, Abagyan, R, Hyvonen, M. | | Deposit date: | 2018-03-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of holoenzyme-disrupting chemicals as substrate-selective CK2 inhibitors.
Sci Rep, 9, 2019
|
|
5VCU
 
 | |
