7RLJ
 
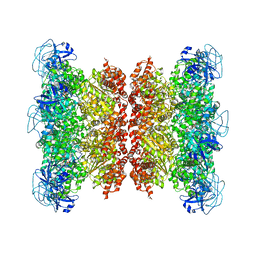 | | Cryo-EM structure of human p97 bound to CB-5083 and ATPgS. | | Descriptor: | 1-[4-(benzylamino)-7,8-dihydro-5H-pyrano[4,3-d]pyrimidin-2-yl]-2-methyl-1H-indole-4-carboxamide, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RL6
 
 | | Cryo-EM structure of human p97-R155H mutant bound to ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLF
 
 | | Cryo-EM structure of human p97-E470D mutant bound to ATPgS. | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLI
 
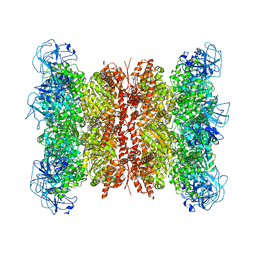 | | Cryo-EM structure of human p97 bound to CB-5083 and ADP. | | Descriptor: | 1-[4-(benzylamino)-7,8-dihydro-5H-pyrano[4,3-d]pyrimidin-2-yl]-2-methyl-1H-indole-4-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLA
 
 | | Cryo-EM structure of human p97-R191Q mutant bound to ATPgS. | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLH
 
 | | Cryo-EM structure of human p97-D592N mutant bound to ATPgS. | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLD
 
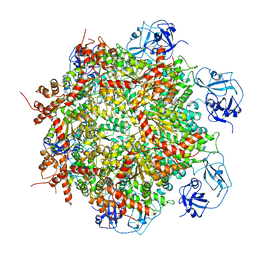 | | Cryo-EM structure of human p97-E470D mutant bound to ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLG
 
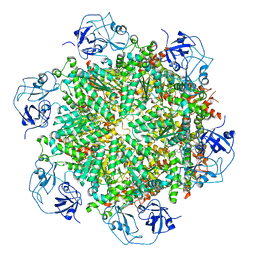 | | Cryo-EM structure of human p97-D592N mutant bound to ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7RLB
 
 | | Cryo-EM structure of human p97-A232E mutant bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Caffrey, B, Zhu, X, Berezuk, A, Tuttle, K, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-22 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation.
J.Biol.Chem., 297, 2021
|
|
7STB
 
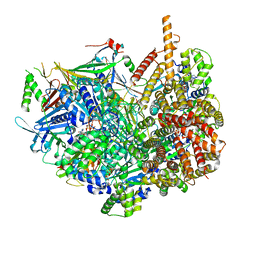 | | Closed state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SH2
 
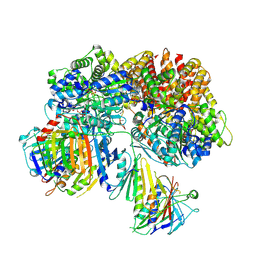 | | Structure of the yeast Rad24-RFC loader bound to DNA and the open 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ST9
 
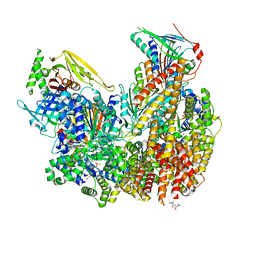 | | Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DNA (5'-D(P*CP*GP*CP*TP*CP*CP*TP*TP*CP*CP*TP*GP*AP*CP*TP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SGZ
 
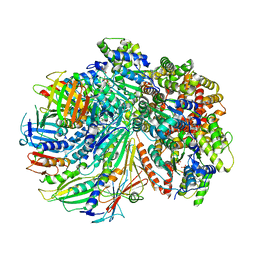 | | Structure of the yeast Rad24-RFC loader bound to DNA and the closed 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7STE
 
 | | Rad24-RFC ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Checkpoint protein RAD24, ... | | Authors: | Castaneda, J.C, Schrecker, M, Remus, D, Hite, R.K. | | Deposit date: | 2021-11-12 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Mechanisms of loading and release of the 9-1-1 checkpoint clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SXO
 
 | | Yeast Lon (PIM1) with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Yang, J, Song, A.S, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2021-11-24 | | Release date: | 2022-01-12 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of hexameric yeast Lon protease (PIM1) highlights the importance of conserved structural elements.
J.Biol.Chem., 298, 2022
|
|
7TKU
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THJ
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TI8
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THV
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIB
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIC
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7T3I
 
 | | CryoEM structure of the Rix7 D2 Walker B mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, Rix7, ... | | Authors: | Kocaman, S, Stanley, R.E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-02 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7T0V
 
 | | CryoEM structure of the crosslinked Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-02 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7SWL
 
 | | CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-03-02 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
