6IHG
 
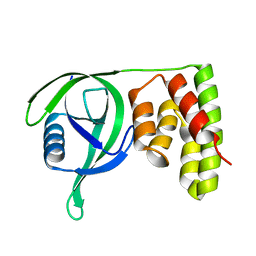 | | N terminal domain of Mycobacterium avium complex Lon protease | | Descriptor: | Lon protease | | Authors: | Chen, X.Y, Zhang, S.J, Bi, F.K, Guo, C.Y, Yao, H.W, Lin, D.H. | | Deposit date: | 2018-09-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
2ANE
 
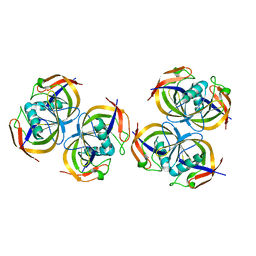 | | Crystal structure of N-terminal domain of E.Coli Lon Protease | | Descriptor: | ATP-dependent protease La | | Authors: | Li, M, Rasulova, F, Melnikov, E.E, Rotanova, T.V, Gustchina, A, Maurizi, M.R, Wlodawer, A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the N-terminal domain of E. coli Lon protease.
Protein Sci., 14, 2005
|
|
3LJC
 
 | | Crystal structure of Lon N-terminal domain. | | Descriptor: | ATP-dependent protease La | | Authors: | Li, M, Gustchina, A, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-01-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the N-terminal fragment of Escherichia coli Lon protease
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1ZBO
 
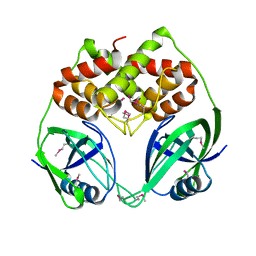 | | X-Ray Crystal Structure of Protein BPP1347 from Bordetella parapertussis. Northeast Structural Genomics Consortium Target BoR27. | | Descriptor: | hypothetical protein BPP1347 | | Authors: | Forouhar, F, Yong, W, Conover, K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Hypothetical Protein BPP1347 from Bordetella parapertussis, Northeast Structural Genomics Target BoR27.
To be Published
|
|
3M65
 
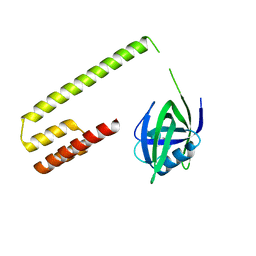 | |
6VBK
 
 | | Crystal structure of N-terminal domain of Mycobacterium tuberculosis complex Lon protease | | Descriptor: | GLYCEROL, Lon211 | | Authors: | Bi, F.K, Chen, C, Chen, X.Y, Guo, C.Y, Lin, D.H. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
7CR9
 
 | |
7CAY
 
 | | Crystal Structure of Lon N-terminal domain protein from Xanthomonas campestris | | Descriptor: | ATP-dependent protease | | Authors: | Singh, R, Sharma, B, Deshmukh, S, Kumar, A, Makde, R.D. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XCC3289 from Xanthomonas campestris: homology with the N-terminal substrate-binding domain of Lon peptidase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7CRA
 
 | |
9DJX
 
 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue WIZ-6 | | Descriptor: | (3S)-3-(5-{[(3R,6S)-1-ethyl-6-methylpiperidin-3-yl]oxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, DNA damage-binding protein 1, Protein Wiz, ... | | Authors: | Partridge, J.R, Ma, X, Ornelas, E. | | Deposit date: | 2024-09-07 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Discovery and Optimization of First-in-Class Molecular Glue Degraders of the WIZ Transcription Factor for Fetal Hemoglobin Induction to Treat Sickle Cell Disease.
J.Med.Chem., 67, 2024
|
|
5V3O
 
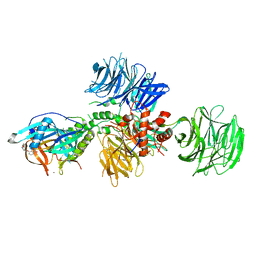 | | Cereblon in complex with DDB1 and CC-220 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Matyskiela, M, Pagarigan, B, Chamberlain, P. | | Deposit date: | 2017-03-07 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Cereblon Modulator (CC-220) with Improved Degradation of Ikaros and Aiolos.
J. Med. Chem., 61, 2018
|
|
4TZ4
 
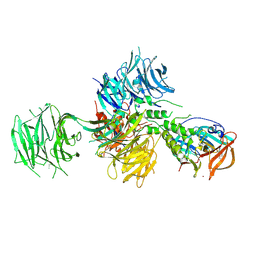 | | Crystal Structure of Human Cereblon in Complex with DDB1 and Lenalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Lenalidomide, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B, Riley, M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
To Be Published
|
|
4CI2
 
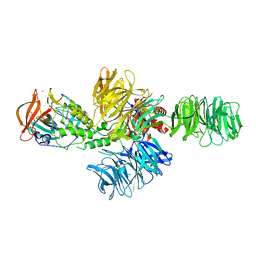 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to lenalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Lenalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
6XK9
 
 | | Cereblon in complex with DDB1, CC-90009, and GSPT1 | | Descriptor: | 2-(4-chlorophenyl)-N-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)-2,2-difluoroacetamide, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | Authors: | Clayton, T.L, Tran, E.T, Zhu, J, Pagarigan, B.E, Matyskiela, M.E, Chamberlain, P.P. | | Deposit date: | 2020-06-25 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells.
Blood, 137, 2021
|
|
4CI1
 
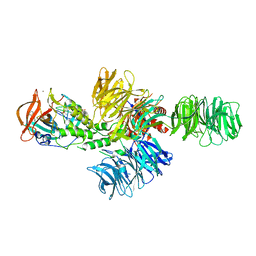 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to thalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Thalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
5HXB
 
 | | Cereblon in complex with DDB1, CC-885, and GSPT1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)urea, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | Authors: | Chamberlain, P.P, Matyskiela, M, Pagarigan, B. | | Deposit date: | 2016-01-30 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase.
Nature, 535, 2016
|
|
4CI3
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
9DJT
 
 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue WIZ-5 | | Descriptor: | (3S)-3-(5-{[(4R)-6-ethyl-6-azaspiro[2.5]octan-4-yl]oxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, DNA damage-binding protein 1, Protein Wiz, ... | | Authors: | Partridge, J.R, Ma, X, Ornelas, E. | | Deposit date: | 2024-09-06 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery and Optimization of First-in-Class Molecular Glue Degraders of the WIZ Transcription Factor for Fetal Hemoglobin Induction to Treat Sickle Cell Disease.
J.Med.Chem., 67, 2024
|
|
9FJX
 
 | |
9DQD
 
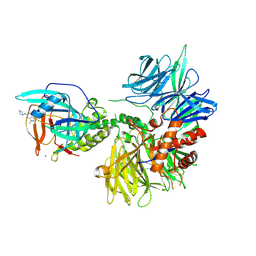 | |
9GAO
 
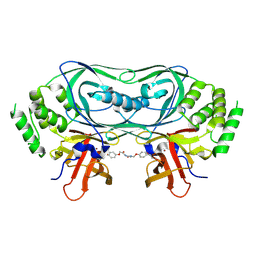 | | Crystal structure of CRBNmidi in complex with 2-(4-(2,6-dioxopiperidin-3-yl)phenoxy)-N-methylacetamide | | Descriptor: | 2-[4-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]phenoxy]-~{N}-methyl-ethanamide, Protein cereblon, ZINC ION | | Authors: | Rutter, Z.J, Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
8RQ1
 
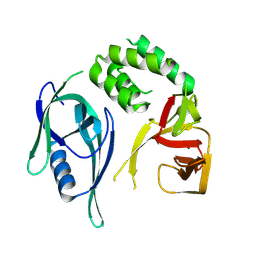 | | Crystal structure of CRBN-midi | | Descriptor: | Protein cereblon, ZINC ION | | Authors: | Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
8RQA
 
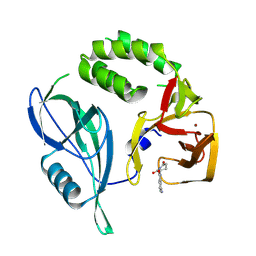 | | Crystal structure of CRBN-midi in complex with Lenalidomide | | Descriptor: | Protein cereblon, S-Lenalidomide, ZINC ION | | Authors: | Rutter, Z.J, Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
8RQ8
 
 | | Crystal structure of CRBN-midi in complex with mezigdomide | | Descriptor: | Mezigdomide, Protein cereblon, ZINC ION | | Authors: | Zollman, D, Kroupova, A, Pethe, J, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
8OIZ
 
 | | Crystal structure of human CRBN-DDB1 in complex with Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Le Bihan, Y.-V, Cabry, M.P, van Montfort, R.L.M. | | Deposit date: | 2023-03-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Chembiochem, 24, 2023
|
|
