5XME
 
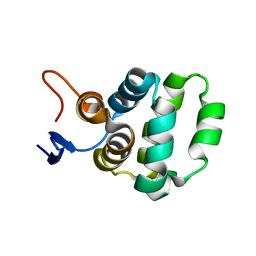 | | Solution structure of C-terminal domain of TRADD | | Descriptor: | Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2017-05-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain of TRADD reveals a novel fold in the death domain superfamily.
Sci Rep, 7, 2017
|
|
1NGR
 
 | |
7CSQ
 
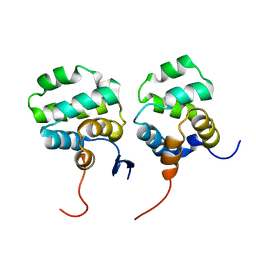 | | Solution structure of the complex between p75NTR-DD and TRADD-DD | | Descriptor: | Tumor necrosis factor receptor superfamily member 16, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Lin, Z, Zhang, N. | | Deposit date: | 2020-08-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of NF-kappa B signaling by the p75 neurotrophin receptor interaction with adaptor protein TRADD through their respective death domains.
J.Biol.Chem., 297, 2021
|
|
5UKE
 
 | |
3OQ9
 
 | | Structure of the FAS/FADD death domain assembly | | Descriptor: | Protein FADD, Tumor necrosis factor receptor superfamily member 6 | | Authors: | Kabaleeswaran, V, Wu, H. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6.8 Å) | | Cite: | The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3EZQ
 
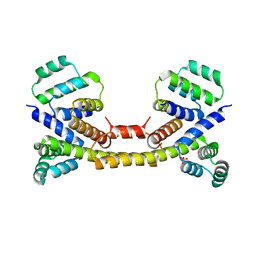 | | Crystal Structure of the Fas/FADD Death Domain Complex | | Descriptor: | Protein FADD, SODIUM ION, SULFATE ION, ... | | Authors: | Schwarzenbacher, R, Robinson, H, Stec, B, Riedl, S.J. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Fas-FADD death domain complex structure unravels signalling by receptor clustering
Nature, 457, 2009
|
|
4O6X
 
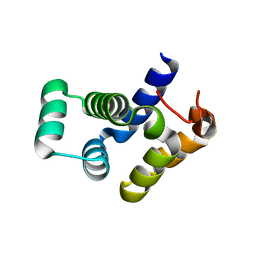 | |
3MOP
 
 | | The ternary Death Domain complex of MyD88, IRAK4, and IRAK2 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, Interleukin-1 receptor-associated kinase-like 2, Myeloid differentiation primary response protein MyD88 | | Authors: | Lin, S.-C, Lo, Y.-C, Wu, H. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling.
Nature, 465, 2010
|
|
4F42
 
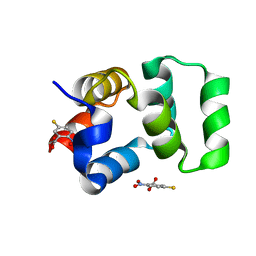 | | Neurotrophin p75NTR intracellular domain | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Qu, Q, Jiang, T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural insights into the allosteric activation mechanism of p75NTR
To be Published
|
|
4F44
 
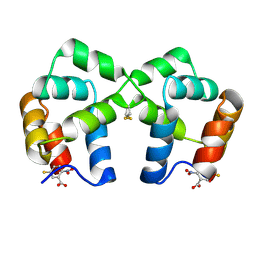 | | Neurotrophin p75NTR intracellular domain | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Qu, Q, Jiang, T. | | Deposit date: | 2012-05-10 | | Release date: | 2013-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the allosteric activation mechanism of p75NTR
To be Published
|
|
2O71
 
 | | Crystal structure of RAIDD DD | | Descriptor: | Death domain-containing protein CRADD | | Authors: | Wu, H, Park, H. | | Deposit date: | 2006-12-09 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of RAIDD Death Domain Implicates Potential Mechanism of PIDDosome Assembly
J.Mol.Biol., 357, 2006
|
|
1ICH
 
 | | SOLUTION STRUCTURE OF THE TUMOR NECROSIS FACTOR RECEPTOR-1 DEATH DOMAIN | | Descriptor: | TUMOR NECROSIS FACTOR RECEPTOR-1 | | Authors: | Sukits, S.F, Lin, L.-L, Malakian, K, Powers, R, Xu, G.-Y. | | Deposit date: | 2001-04-01 | | Release date: | 2002-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tumor necrosis factor receptor-1 death domain.
J.Mol.Biol., 310, 2001
|
|
2OF5
 
 | | Oligomeric Death Domain complex | | Descriptor: | Death domain-containing protein CRADD, Leucine-rich repeat and death domain-containing protein | | Authors: | Park, H.H, Logette, E, Raunser, S, Cuenin, S, Walz, T, Tschopp, J, Wu, H. | | Deposit date: | 2007-01-02 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex.
Cell(Cambridge,Mass.), 128, 2007
|
|
6HJ7
 
 | |
6I3N
 
 | |
2YQF
 
 | | Solution structure of the death domain of Ankyrin-1 | | Descriptor: | Ankyrin-1 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the death domain of Ankyrin-1
To be Published
|
|
2YVI
 
 | | Crystal structure of a death domain of human ankryn protein | | Descriptor: | Ankyrin-1, GLYCEROL | | Authors: | Ihsanawati, Bessho, Y, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-12 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of a death domain of human ankryn protein
To be Published
|
|
1IK7
 
 | | Crystal Structure of the Uncomplexed Pelle Death Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PROBABLE SERINE/THREONINE-PROTEIN KINASE Pelle | | Authors: | Xiao, T, Gardner, K.H, Sprang, S.R. | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cosolvent-induced transformation
of a death domain tertiary structure
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1WXP
 
 | | Solution structure of the death domain of nuclear matrix protein p84 | | Descriptor: | THO complex subunit 1 | | Authors: | Niraula, T.N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-28 | | Release date: | 2005-07-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the death domain of nuclear matrix protein p84
To be Published
|
|
1WMG
 
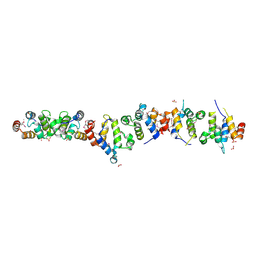 | | Crystal structure of the UNC5H2 death domain | | Descriptor: | SULFATE ION, SULFITE ION, netrin receptor Unc5h2 | | Authors: | Handa, N, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-09 | | Release date: | 2005-01-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the UNC5H2 death domain
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2N97
 
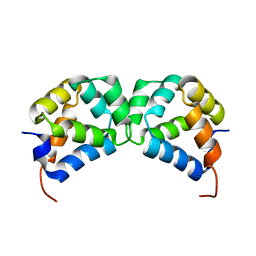 | | DD homodimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 16 | | Authors: | Lin, Z, Ibanez, C.F. | | Deposit date: | 2015-11-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of death domain signaling in the p75 neurotrophin receptor
Elife, 4, 2015
|
|
1YGO
 
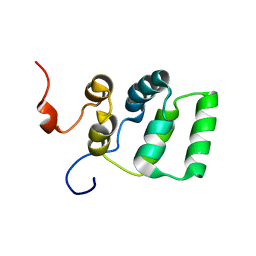 | |
2DBF
 
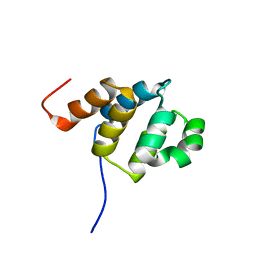 | | Solution structure of the Death domain in human Nuclear factor NF-kappa-B p105 subunit | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Saito, K, Inoue, M, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Death domain in human Nuclear factor NF-kappa-B p105 subunit
To be Published
|
|
2D96
 
 | |
2IB1
 
 | |
