2NBW
 
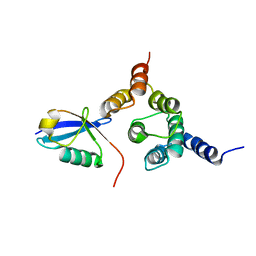 | | Solution structure of the Rpn1 T1 site with the Rad23 UBL domain | | Descriptor: | 26S proteasome regulatory subunit RPN1, UV excision repair protein RAD23 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
7RBR
 
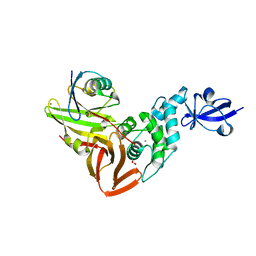 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Papain-like protease, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
2NBV
 
 | |
6UKU
 
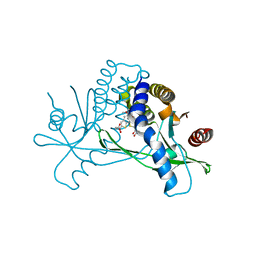 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 3 | | Descriptor: | 4,4'-[propane-1,3-diylbis(6-methoxy-1-benzothiene-5,2-diyl)]bis(4-oxobutanoic acid), fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
6UL0
 
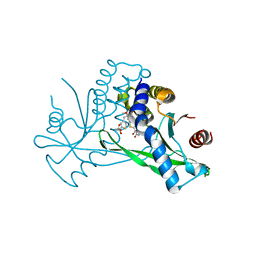 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 4 | | Descriptor: | 4-{5-[(1Z)-3-{[2-(3-carboxypropanoyl)-6-methoxy-1-benzothiophen-5-yl]oxy}prop-1-en-1-yl]-6-methoxy-1-benzothiophen-2-yl}-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
6UKV
 
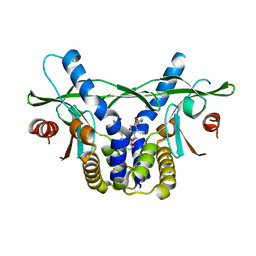 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 9 | | Descriptor: | 4-[6-(3-{[2-(3-carboxypropanoyl)-6-methoxy-1-benzothiophen-5-yl]oxy}propoxy)-5-methoxy-1-benzothiophen-2-yl]-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
7R70
 
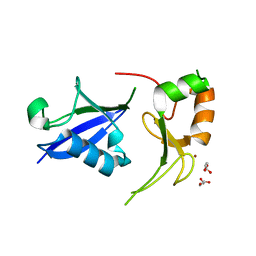 | | Crystal Structure of the UbArk2C fusion protein | | Descriptor: | GLYCEROL, Ubiquitin,E3 ubiquitin-protein ligase RNF165, ZINC ION | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
5C1Z
 
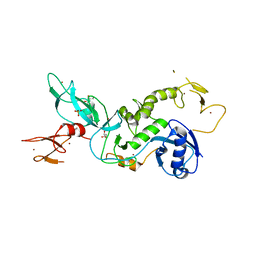 | | Parkin (UblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
5CHV
 
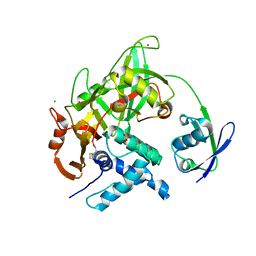 | | Crystal structure of USP18-ISG15 complex | | Descriptor: | CHLORIDE ION, SULFATE ION, Ubiquitin-like protein ISG15, ... | | Authors: | Fritz, G, Basters, A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural basis of the specificity of USP18 toward ISG15.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5C7J
 
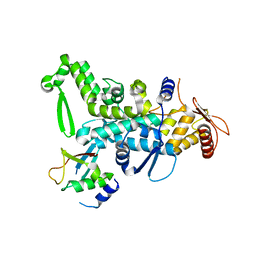 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
6UYJ
 
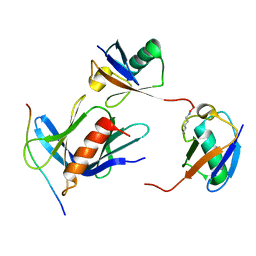 | | hRpn13:hRpn2:K48-diubiquitin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-20 | | Method: | SOLUTION NMR | | Cite: | An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure.
Structure, 28, 2020
|
|
6VAF
 
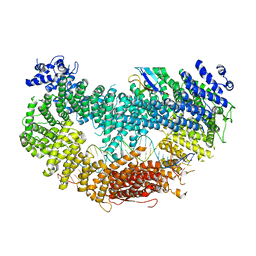 | |
6VAE
 
 | |
6UPU
 
 | |
6UYI
 
 | | hRpn13:hRpn2:K48-diubiquitin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure.
Structure, 28, 2020
|
|
6TUV
 
 | |
4ZYN
 
 | | Crystal Structure of Parkin E3 ubiquitin ligase (linker deletion; delta 86-130) | | Descriptor: | E3 ubiquitin-protein ligase parkin, SULFATE ION, ZINC ION | | Authors: | Lilov, A, Sauve, V, Trempe, J.F, Rodionov, D, Wang, J, Gehring, K. | | Deposit date: | 2015-05-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A Ubl/ubiquitin switch in the activation of Parkin.
Embo J., 34, 2015
|
|
7Z77
 
 | | Crystal structure of compound 6 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{S})-4-[4-(4-bromanyl-7-cyclopentyl-5-oxidanylidene-benzimidazolo[1,2-a]quinazolin-9-yl)piperidin-1-yl]-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]butyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
5AF6
 
 | | Structure of Lys33-linked diUb bound to Trabid NZF1 | | Descriptor: | TRABID, UBIQUITIN, ZINC ION | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
7Z76
 
 | | Crystal structure of compound 10 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[(1~{R})-2-[(2~{R})-1-[4-(4-bromanyl-7-cyclopentyl-5-oxidanylidene-benzimidazolo[1,2-a]quinazolin-9-yl)piperidin-1-yl]propan-2-yl]oxy-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-1-[(2~{S})-2-[[1-(dimethylamino)cyclopropyl]carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
7SDE
 
 | | Cryo-EM structure of Nse5/6 heterodimer | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-09-29 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of Nse5/6 complex with the C terminal part of Nse5
To Be Published
|
|
5CAW
 
 | |
7SHL
 
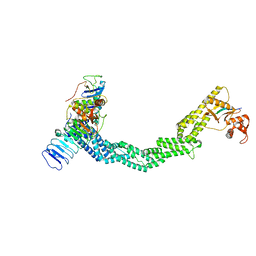 | | Structure of Xenopus laevis CRL2Lrr1 (State 2) | | Descriptor: | CULLIN_2 domain-containing protein, Elongin-C, Lrr1, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2021-10-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of CRL2Lrr1, the E3 ubiquitin ligase that promotes DNA replication termination in vertebrates.
Nucleic Acids Res., 49, 2021
|
|
7SHK
 
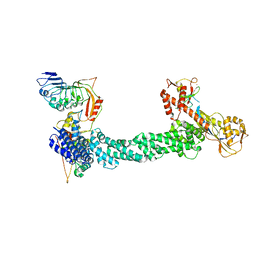 | | Structure of Xenopus laevis CRL2Lrr1 (State 1) | | Descriptor: | CULLIN_2 domain-containing protein, Elongin-C, Lrr1, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2021-10-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of CRL2Lrr1, the E3 ubiquitin ligase that promotes DNA replication termination in vertebrates.
Nucleic Acids Res., 49, 2021
|
|
7RUC
 
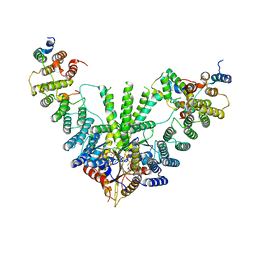 | | Metazoan pre-targeting GET complex with SGTA (cBUGGS) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 4 homolog, ... | | Authors: | Keszei, A.F.A, Yip, M.C.J, Shao, S. | | Deposit date: | 2021-08-16 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into metazoan pretargeting GET complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
