6QNX
 
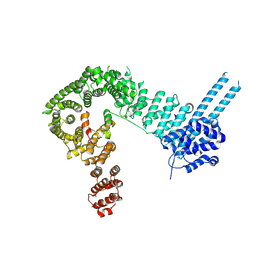 | | Structure of the SA2/SCC1/CTCF complex | | Descriptor: | Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog, Transcriptional repressor CTCF | | Authors: | Li, Y, Muir, K.W, Panne, D. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for cohesin-CTCF-anchored loops.
Nature, 578, 2020
|
|
6UML
 
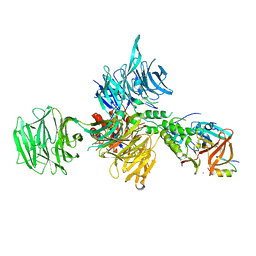 | | Structural Basis for Thalidomide Teratogenicity Revealed by the Cereblon-DDB1-SALL4-Pomalidomide Complex | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clayton, T.L, Matyskiela, M.E, Pagarigan, B.E, Tran, E.T, Chamberlain, P.P. | | Deposit date: | 2019-10-09 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Crystal structure of the SALL4-pomalidomide-cereblon-DDB1 complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1UN6
 
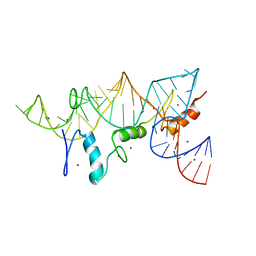 | | THE CRYSTAL STRUCTURE OF A ZINC FINGER - RNA COMPLEX REVEALS TWO MODES OF MOLECULAR RECOGNITION | | Descriptor: | 5S RIBOSOMAL RNA, MAGNESIUM ION, TRANSCRIPTION FACTOR IIIA, ... | | Authors: | Lu, D, Searles, M.A, Klug, A. | | Deposit date: | 2003-09-04 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Zinc-Finger-RNA Complex Reveals Two Modes of Molecular Recognition
Nature, 426, 2003
|
|
2RV6
 
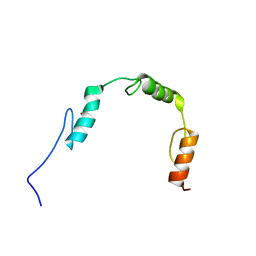 | | Solution structures of the DNA-binding domains (ZF2-ZF3-ZF4) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RSJ
 
 | |
2RPC
 
 | |
8D7Z
 
 | | Cereblon-DDB1 bound to CC-92480 and Ikaros ZF1-2-3 | | Descriptor: | DNA damage-binding protein 1, DNA-binding protein Ikaros, Mezigdomide, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8D80
 
 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8DEY
 
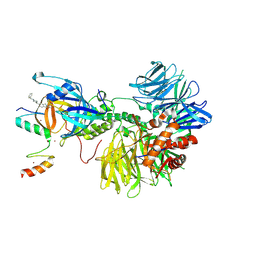 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
1TF6
 
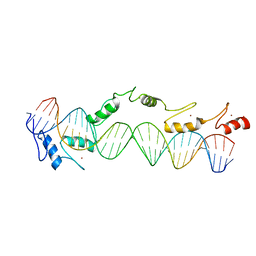 | | CO-CRYSTAL STRUCTURE OF XENOPUS TFIIIA ZINC FINGER DOMAIN BOUND TO THE 5S RIBOSOMAL RNA GENE INTERNAL CONTROL REGION | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*GP*CP*CP*TP*GP*GP*TP*TP*AP*GP*TP*AP*C P*CP*TP*GP*GP*AP* TP*GP*GP*GP*AP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*TP*CP*CP*CP*AP*TP*CP*CP*AP*GP*GP*T P*AP*CP*TP*AP*AP* CP*CP*AP*GP*GP*CP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR IIIA), ... | | Authors: | Nolte, R.T, Conlin, R.M, Harrison, S.C, Brown, R.S. | | Deposit date: | 1998-03-02 | | Release date: | 1998-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
8TNR
 
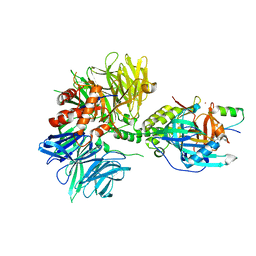 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8TNP
 
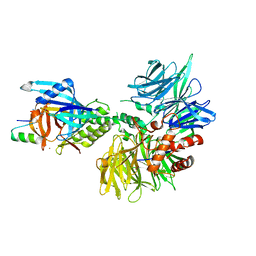 | | Cryo-EM structure of DDB1dB:CRBN:Pomalidomide:SD40 | | Descriptor: | DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, Protein cereblon, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
8TNQ
 
 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 1 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
2LVU
 
 | | Solution structure of Miz-1 zinc finger 10 | | Descriptor: | ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Bedard, M, Maltais, L, Beaulieu, M, Bernard, D, Lavigne, P. | | Deposit date: | 2012-07-11 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: solution structure of human Miz-1 zinc fingers 8 to 10.
J.Biomol.Nmr, 54, 2012
|
|
2LVR
 
 | | Solution structure of Miz-1 zinc finger 8 | | Descriptor: | ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Bedard, M, Maltais, L, Beaulieu, M, Bernard, D, Lavigne, P. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: solution structure of human Miz-1 zinc fingers 8 to 10.
J.Biomol.Nmr, 54, 2012
|
|
2LVT
 
 | | Solution structure of Miz-1 zinc finger 9 | | Descriptor: | ZINC ION, Zinc finger and BTB domain-containing protein 17 | | Authors: | Bedard, M, Maltais, L, Beaulieu, M, Bernard, D, Lavigne, P. | | Deposit date: | 2012-07-11 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: solution structure of human Miz-1 zinc fingers 8 to 10.
J.Biomol.Nmr, 54, 2012
|
|
2M0F
 
 | |
2M0E
 
 | |
2M0D
 
 | |
2HGH
 
 | |
2J7J
 
 | |
