5FTI
 
 | |
5FXH
 
 | | GluN1b-GluN2B NMDA receptor in non-active-1 conformation | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5EHM
 
 | |
5ELV
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L504-N775) in complex with glutamate and BPAM-521 at 1.92 A resolution | | Descriptor: | 4-Cyclopropyl-3,4-dihydro-7-hydroxy-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Krintel, C, Juknaite, L, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-11-05 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Enthalpy-Entropy Compensation in the Binding of Modulators at Ionotropic Glutamate Receptor GluA2.
Biophys.J., 110, 2016
|
|
5FHN
 
 | |
3B7D
 
 | |
5EHS
 
 | |
3BFU
 
 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | Descriptor: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
3B6Q
 
 | |
3B6W
 
 | |
3DP6
 
 | | Crystal structure of the binding domain of the AMPA subunit GluR2 bound to glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | Deposit date: | 2008-07-07 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
3EN3
 
 | | Crystal Structure of the GluR4 Ligand-Binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 4,Glutamate receptor | | Authors: | Gill, A, Madden, D.R. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-19 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Correlating AMPA receptor activation and cleft closure across subunits: crystal structures of the GluR4 ligand-binding domain in complex with full and partial agonists
Biochemistry, 47, 2008
|
|
3C36
 
 | |
3C34
 
 | |
5FXG
 
 | | GLUN1B-GLUN2B NMDA RECEPTOR IN ACTIVE CONFORMATION | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
3EPE
 
 | |
5FXJ
 
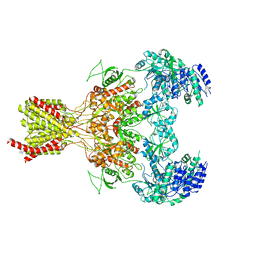 | | GluN1b-GluN2B NMDA receptor structure-Class X | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, NMDA 1, NMDA 2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa H, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
3DP4
 
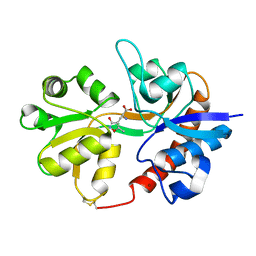 | | Crystal structure of the binding domain of the AMPA subunit GluR3 bound to AMPA | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | Deposit date: | 2008-07-07 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
3C33
 
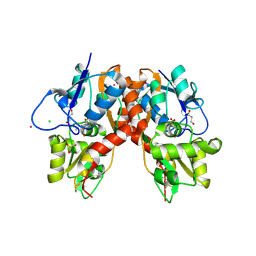 | |
5H8H
 
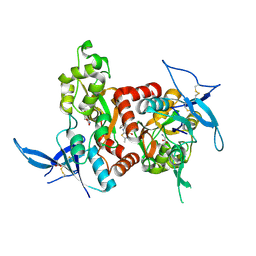 | | Structure of the human GluN1/GluN2A LBD in complex with GNE3419 | | Descriptor: | 7-[[ethyl(phenyl)amino]methyl]-2-methyl-[1,3,4]thiadiazolo[3,2-a]pyrimidin-5-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
5H8F
 
 | | Structure of the apo human GluN1/GluN2A LBD | | Descriptor: | GLUTAMIC ACID, GLYCEROL, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
5H8S
 
 | | Structure of the human GluA2 LBD in complex with GNE3419 | | Descriptor: | 7-[[ethyl(phenyl)amino]methyl]-2-methyl-[1,3,4]thiadiazolo[3,2-a]pyrimidin-5-one, CACODYLATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
3C32
 
 | |
5H8Q
 
 | | Structure of the human GluN1/GluN2A LBD in complex with GNE8324 | | Descriptor: | 6-[[ethyl-(4-fluorophenyl)amino]methyl]-2,3-dihydro-1~{H}-cyclopenta[3,4][1,3]thiazolo[1,4-~{a}]pyrimidin-8-one, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
3BKI
 
 | | Crystal Structure of the GluR2 ligand binding core (S1S2J) in complex with FQX at 1.87 Angstroms | | Descriptor: | Glutamate receptor 2, [1,2,5]oxadiazolo[3,4-g]quinoxaline-6,7(5H,8H)-dione 1-oxide | | Authors: | Cruz, L, Estebanez-Perpina, E, Pfaff, S, Borngraeber, S, Bao, N, Fletterick, R, England, P. | | Deposit date: | 2007-12-06 | | Release date: | 2008-09-16 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | 6-Azido-7-nitro-1,4-dihydroquinoxaline-2,3-dione (ANQX) forms an irreversible bond to the active site of the GluR2 AMPA receptor.
J.Med.Chem., 51, 2008
|
|
