7MW7
 
 | | Crystal structure of P1G mutant of D-dopachrome tautomerase | | Descriptor: | D-dopachrome decarboxylase, SODIUM ION, SULFATE ION | | Authors: | Manjula, R, Murphy, E.L, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-15 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
3KAN
 
 | |
8VFL
 
 | | Crystal Structure of WT D-Dopachrome Tautomerase (D-DT) at 290K | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG7
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
7MSE
 
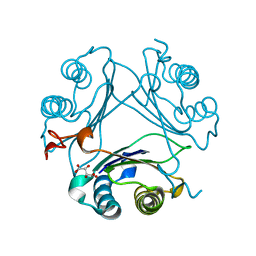 | | High-resolution crystal structure of hMIF2 with tartrate at the active site | | Descriptor: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
8VFN
 
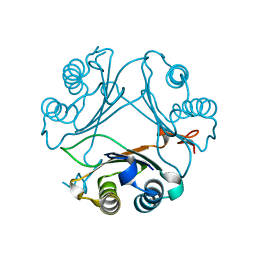 | | Crystal Structure of WT D-Dopachrome Tautomerase (D-DT) at 310K | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8DBB
 
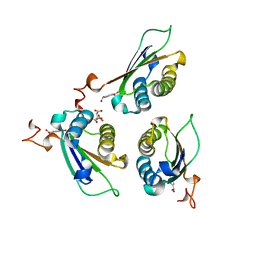 | | Crystal structure of DDT with the selective inhibitor 2,5-Pyridinedicarboxylic Acid | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase, pyridine-2,5-dicarboxylic acid | | Authors: | Parkins, A, Banumathi, S, Pantouris, G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 2,5-Pyridinedicarboxylic acid is a bioactive and highly selective inhibitor of D-dopachrome tautomerase.
Structure, 31, 2023
|
|
8VFW
 
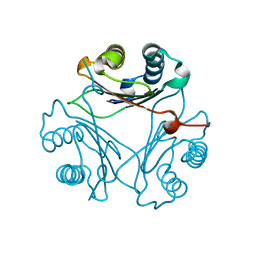 | | Crystal Structure of V113N D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG8
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) Bound to 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
7MRU
 
 | | Crystal structure of S62A MIF2 mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-dopachrome decarboxylase | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
8VFO
 
 | | Crystal Structure of L117G Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
6C5F
 
 | |
8VG5
 
 | | Crystal Structure of V113N Variant of D-Dopachrome Tautomerase (D-DT) Bound with 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
1DPT
 
 | | D-DOPACHROME TAUTOMERASE | | Descriptor: | D-DOPACHROME TAUTOMERASE | | Authors: | Sugimoto, H, Taniguchi, M, Nakagawa, A, Tanaka, I. | | Deposit date: | 1998-05-11 | | Release date: | 1999-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of human D-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 A resolution.
Biochemistry, 38, 1999
|
|
7MRV
 
 | | F100A mutant structure of MIF2 (D-DT) | | Descriptor: | D-dopachrome decarboxylase, SULFATE ION | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
8VFK
 
 | | Crystal Structure of Delta 109-117 D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRATE ANION, D-dopachrome decarboxylase, SODIUM ION | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
4Q3F
 
 | | Human D-DT complexed with tartrate | | Descriptor: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | Authors: | Rajasekaran, D, Lolis, E. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting distinct tautomerase sites of D-DT and MIF with a single molecule for inhibition of neutrophil lung recruitment.
Faseb J., 28, 2014
|
|
8VDY
 
 | | Crystal Structure of Delta 114-117 D-Dopachrome Tautomerase (D-DT) | | Descriptor: | D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
