6IH0
 
 | | Aquifex aeolicus LpxC complex with ACHN-975 | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Li, D.Y, Fan, S, Jin, Y.Y, Zhang, C, Lv, G.X, Wu, G.T, Yang, Z.Y. | | Deposit date: | 2018-09-27 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Complex Structure of Protein AaLpxC from Aquifex aeolicus with ACHN-975 Molecule Suggests an Inhibitory Mechanism at Atomic-Level against Gram-Negative Bacteria
Molecules, 26, 2021
|
|
4U3D
 
 | |
3P3C
 
 | | Crystal Structure of the Aquifex aeolicus LpxC/LPC-009 complex | | Descriptor: | N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, PHOSPHATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design.
Chem.Biol., 18, 2011
|
|
4U3B
 
 | |
4OZE
 
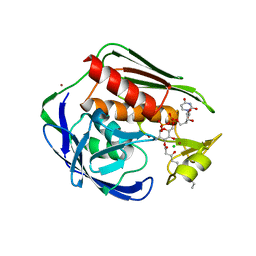 | | A.aolicus LpxC in complex with native product | | Descriptor: | CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Olivier, N.B. | | Deposit date: | 2014-02-15 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mechanistic insight from the crystal structure of A.aolicus LpxC in the presence of product
To Be Published
|
|
5U86
 
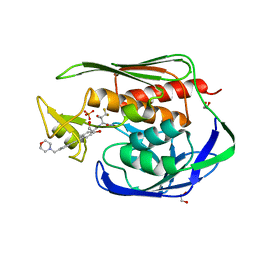 | | Structure of the Aquifex aeolicus LpxC/LPC-069 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-({4-[(morpholin-4-yl)methyl]phenyl}ethynyl)benzamide, ... | | Authors: | Najeeb, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2016-12-13 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Curative Treatment of Severe Gram-Negative Bacterial Infections by a New Class of Antibiotics Targeting LpxC.
MBio, 8, 2017
|
|
5DRP
 
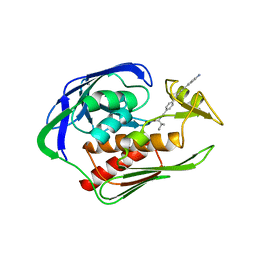 | | Structure of the AaLpxC/LPC-023 Complex | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N~2~-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-isoleucinamide, ... | | Authors: | Najeeb, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
3P76
 
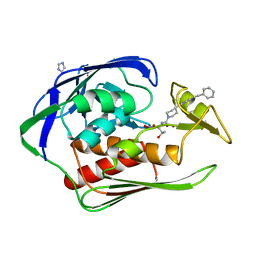 | | X-ray crystal structure of Aquifex aeolicus LpxC complexed SCH1379777 | | Descriptor: | IMIDAZOLE, N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-[4-(phenylethynyl)phenyl]piperidine-1-carboxamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Orth, P. | | Deposit date: | 2010-10-12 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and synthesis of potent Gram-negative specific LpxC inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1P42
 
 | | Crystal structure of Aquifex aeolicus LpxC Deacetylase (Zinc-Inhibited Form) | | Descriptor: | MYRISTIC ACID, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Whittington, D.A, Rusche, K.M, Shin, H, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of LpxC, a Zinc-Dependent Deacetylase Essential for Endotoxin Biosynthesis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2GO3
 
 | | Crystal structure of Aquifex aeolicus LpxC complexed with imidazole. | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
5DRO
 
 | | Structure of the Aquifex aeolicus LpxC/LPC-011 Complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
1YHC
 
 | | Crystal structure of Aquifex aeolicus LpxC deacetylase complexed with cacodylate | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hernick, M, Gennadios, H.A, Whittington, D.A, Rusche, K.M, Christianson, D.W, Fierke, C.A. | | Deposit date: | 2005-01-07 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | UDP-3-O-((R)-3-hydroxymyristoyl)-N-acetylglucosamine Deacetylase Functions through a General Acid-Base Catalyst Pair Mechanism
J.Biol.Chem., 280, 2005
|
|
2J65
 
 | | Structure of LpxC from Aquifex aeolicus in complex with UDP | | Descriptor: | CHLORIDE ION, MYRISTIC ACID, UDP-3-O-[3-HYDROXYMYRISTOYL] N-ACETYLGLUCOSAMINE DEACETYLASE, ... | | Authors: | Buetow, L, Dawson, A, Hunter, W.N. | | Deposit date: | 2006-09-26 | | Release date: | 2006-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Nucleotide-Binding Site of Aquifex Aeolicus Lpxc.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2O3Z
 
 | | X-ray crystal structure of LpxC complexed with 3-heptyloxybenzoate | | Descriptor: | 3-(heptyloxy)benzoic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Christianson, D.W. | | Deposit date: | 2006-12-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Amphipathic benzoic acid derivatives: synthesis and binding in the hydrophobic tunnel of the zinc deacetylase LpxC.
Bioorg.Med.Chem., 15, 2007
|
|
2IER
 
 | |
2GO4
 
 | | Crystal structure of Aquifex aeolicus LpxC complexed with TU-514 | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
1YH8
 
 | | Crystal structure of Aquifex aeolicus LpxC deacetylase complexed with palmitate | | Descriptor: | CHLORIDE ION, PALMITOLEIC ACID, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Hernick, M, Gennadios, H.A, Whittington, D.A, Rusche, K.M, Christianson, D.W, Fierke, C.A. | | Deposit date: | 2005-01-07 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | UDP-3-O-((R)-3-hydroxymyristoyl)-N-acetylglucosamine Deacetylase Functions through a General Acid-Base Catalyst Pair Mechanism
J.Biol.Chem., 280, 2005
|
|
2IES
 
 | |
1XXE
 
 | | RDC refined solution structure of the AaLpxC/TU-514 complex | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Coggins, B.E, McClerren, A.L, Jiang, L, Li, X, Rudolph, J, Hindsgaul, O, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the LpxC-TU-514 Complex and pK(a) Analysis of an Active Site Histidine: Insights into the Mechanism and Inhibitor Design
Biochemistry, 44, 2005
|
|
2JT2
 
 | | Solution Structure of the Aquifex aeolicus LpxC- CHIR-090 complex | | Descriptor: | N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Barb, A.W, Jiang, L, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2007-07-18 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: Time-dependent inhibition and specificity in ligand binding
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
