8WDO
 
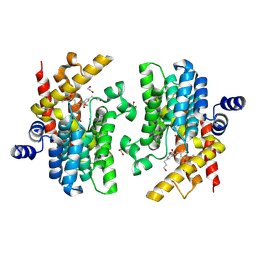 | | Crystal structure of PDE4D complexed with DCN | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-2-(3,4-dimethoxyphenyl)-3-(hydroxymethyl)-7-methoxy-2,3-dihydro-1-benzofuran-5-yl]propan-1-ol, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of Dihydrobenzofuran Neolignans as Novel PDE4 Inhibitors and Evaluation of Antiatopic Dermatitis Efficacy in DNCB-Induced Mice Model.
J.Med.Chem., 67, 2024
|
|
8Z6R
 
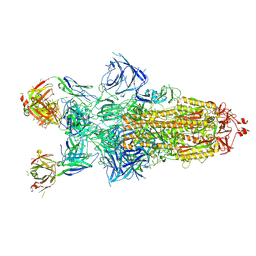 | | Structure of XBB.1.16 S trimer with 3 down-RBDs complex with antibody CYFN1006-1. | | Descriptor: | CYFN1006-1 heavy chain, CYFN1006-1 light chain, Spike glycoprotein,Fibritin,Spike glycoprotein,Fibritin,Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Sun, L. | | Deposit date: | 2024-04-19 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structure of XBB.1.16 S trimer with 3 down-RBDs complex with antibody CYFN1006-1.
To Be Published
|
|
5GWB
 
 | |
6TEN
 
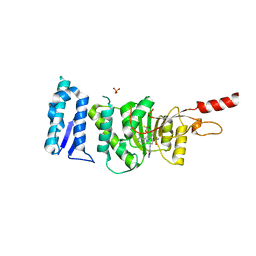 | | Crystal structure of Dot1L in complex with an inhibitor (compound 11). | | Descriptor: | 3-[(4-azanyl-6-methoxy-1,3,5-triazin-2-yl)amino]-4-[[(~{S})-[2,2-bis(fluoranyl)-1,3-benzodioxol-4-yl]-(3-chloranylpyridin-2-yl)methyl]amino]benzenesulfonamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
6OM1
 
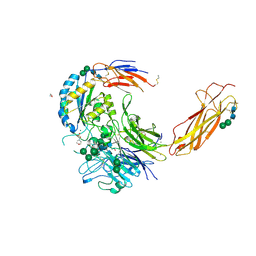 | | Crystal structure of an atypical integrin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.C, Springer, T.A. | | Deposit date: | 2019-04-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | General structural features that regulate integrin affinity revealed by atypical alpha V beta 8.
Nat Commun, 10, 2019
|
|
8Z6T
 
 | |
6I16
 
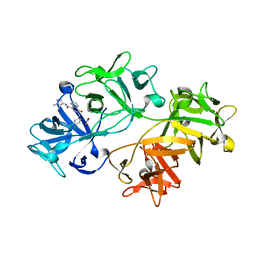 | | CRYSTAL STRUCTURE OF FASCIN IN COMPLEX WITH COMPOUND 15 | | Descriptor: | 6-[(3,4-dichlorophenyl)methyl]-~{N}-(1-methylpyrazol-4-yl)-5-oxidanylidene-1,6-naphthyridine-8-carboxamide, ACETATE ION, Fascin | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2018-10-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis and biological evaluation of a novel series of isoquinolone and pyrazolo[4,3-c]pyridine inhibitors of fascin 1 as potential anti-metastatic agents.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6TFZ
 
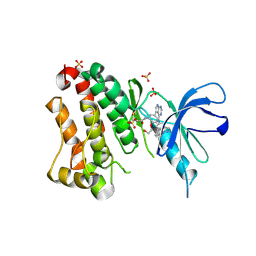 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 19 | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, SULFATE ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
7QSI
 
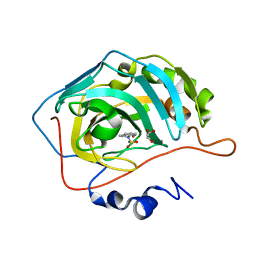 | | human Carbonic Anhydrase II in complex with indoline-1-sulfonamide | | Descriptor: | 2,3-dihydroindole-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-01-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Diversely substituted sulfamides for fragment-based drug discovery of carbonic anhydrase inhibitors: synthesis and inhibitory profile.
J Enzyme Inhib Med Chem, 37, 2022
|
|
8YZR
 
 | |
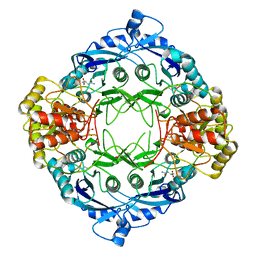
5DND
 
 | |
5H4U
 
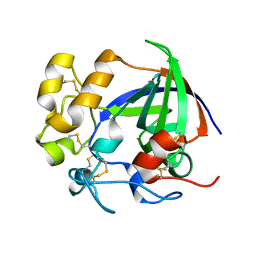 | | Crystal structure of cellulase from Antarctic springtail, Cryptopygus antarcticus | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | An, Y.J, Hong, S.K, Cha, S.S. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic and Structural Characterization of a Thermo-Tolerant, Cold-Active, and Acidic Endo-beta-1,4-glucanase from Antarctic Springtail, Cryptopygus antarcticus.
J. Agric. Food Chem., 65, 2017
|
|
6EFV
 
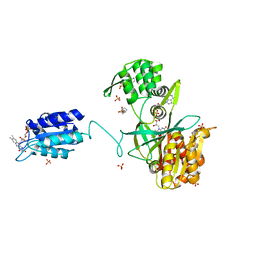 | | The NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation that is unique to this diflavin reductase | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tavolieri, A.M, Askenasy, I, Murray, D.T, Pennington, J.M, Stroupe, M.E. | | Deposit date: | 2018-08-17 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase.
J. Struct. Biol., 205, 2019
|
|
4RER
 
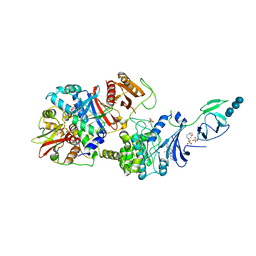 | | Crystal structure of the phosphorylated human alpha1 beta2 gamma1 holo-AMPK complex bound to AMP and cyclodextrin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, ... | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.047 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
9G6I
 
 | |
8Z7M
 
 | | The crystal structure of AFM-1 | | Descriptor: | Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Xiao, Y.J, Wang, Z.F. | | Deposit date: | 2024-04-20 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into the subclass B1 metallo-beta-lactamase AFM-1.
Biochem.Biophys.Res.Commun., 720, 2024
|
|
6Q7H
 
 | |
8I5T
 
 | |
8I5U
 
 | |
5DXV
 
 | | Crystal structure of Rethreaded DHFR | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Faham, S. | | Deposit date: | 2015-09-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Protein rethreading: A novel approach to protein design.
Sci Rep, 6, 2016
|
|
4ZKN
 
 | |
7BU3
 
 | | Structure of alcohol dehydrogenase YjgB in complex with NADP from Escherichia coli | | Descriptor: | ASPARTIC ACID, Alcohol dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nguyen, G.T, Kim, Y.-G, Ahn, J.-W, Chang, J.H. | | Deposit date: | 2020-04-03 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Broad Substrate Selectivity of Alcohol Dehydrogenase YjgB from Escherichia coli .
Molecules, 25, 2020
|
|
5K2O
 
 | | Crystal structure of Arabidopsis thaliana acetohydroxyacid synthase in complex with a pyrimidinyl-benzoate herbicide, pyrithiobac | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2-chloranyl-6-(4,6-dimethoxypyrimidin-2-yl)sulfanyl-benzoic acid, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8A1V
 
 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5U39
 
 | | Pseudomonas aeruginosa LpxC in complex with CHIR-090 | | Descriptor: | N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Sprague, E.R. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC.
J. Med. Chem., 60, 2017
|
|
