8DG6
 
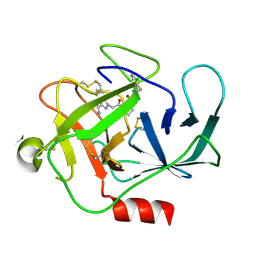 | |
4ZII
 
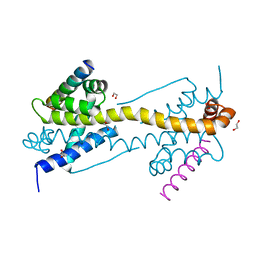 | | Crystal Structure of core/latch dimer of BaxI66A in complex with BidBH3 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, BH3-interacting domain death agonist | | Authors: | Czabotar, P.E, Robin, A.Y, Krishna Kumar, K, Westphal, D, Wardak, A.Z, Thompson, G.V, Dewson, G, Colman, P.M. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.
Cell Death Dis, 6, 2015
|
|
4WK0
 
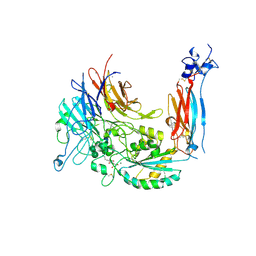 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARG-GLY-ASP, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6EK1
 
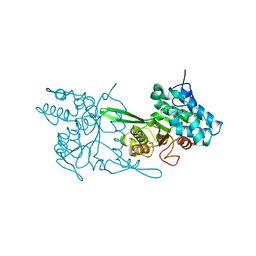 | | Crystal structure of Type IIP restriction endonuclease PfoI | | Descriptor: | 1,2-ETHANEDIOL, restriction endonuclease PfoI | | Authors: | Tamulaitiene, G, Manakova, E, Jovaisaite, V, Grazulis, S, Siksnys, V. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Unique mechanism of target recognition by PfoI restriction endonuclease of the CCGG-family.
Nucleic Acids Res., 47, 2019
|
|
4RIV
 
 | | A Carbonic Anhydrase IX Mimic in Complex with Saccharin | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Mahon, B.P, McKenna, R. | | Deposit date: | 2014-10-07 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Saccharin: A lead compound for structure-based drug design of carbonic anhydrase IX inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
6YL4
 
 | | Soluble epoxide hydrolase in complex with 3-((R)-3-(1-hydroxyureido)but-1-yn-1-yl)-N-((S)-3-phenyl-3-(4-trifluoromethoxy)phenyl)propyl)benzamide | | Descriptor: | 3-[(3~{R})-3-[aminocarbonyl(oxidanyl)amino]but-1-ynyl]-~{N}-[(3~{S})-3-phenyl-3-[4-(trifluoromethyloxy)phenyl]propyl]benzamide, Bifunctional epoxide hydrolase 2 | | Authors: | Kramer, J.S, Pogoryelov, D, Hiesinger, K, Proschak, E. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationship Studies of Dual Inhibitors of Soluble Epoxide Hydrolase and 5-Lipoxygenase.
J.Med.Chem., 63, 2020
|
|
7MGU
 
 | |
5GQJ
 
 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Ser193 and Ala194 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
8WXI
 
 | |
8AOZ
 
 | | ForT Mutant L24A | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-3-[(2~{S},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]-1~{H}-pyrazole-5-carboxylic acid, Beta-ribofuranosylaminobenzene 5'-phosphate synthase, ... | | Authors: | Li, W, Naismith, J.H. | | Deposit date: | 2022-08-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental and computational snapshots of C-C bond formation in a C-nucleoside synthase.
Open Biology, 13, 2023
|
|
8IBQ
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-02-10 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
6W30
 
 | | Protein Tyrosine Phosphatase 1B Bound to Amorphadiene | | Descriptor: | Amorphadiene, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sarkar, A, Kim, E.Y, Hongdusit, A, Sankaran, B, Fox, J. | | Deposit date: | 2020-03-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Microbially Guided Discovery and Biosynthesis of Biologically Active Natural Products.
Acs Synth Biol, 10, 2021
|
|
8WRW
 
 | |
8DQF
 
 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide | | Descriptor: | Carbonic anhydrase, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)cyclohexanecarboxamide, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8DR2
 
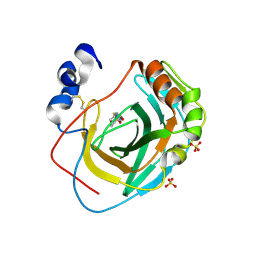 | | Crystal structure of Neisseria gonorrhoeae carbonic anhydrase with 2-cyclohexyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)acetamide | | Descriptor: | 2-cyclohexyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)acetamide, Carbonic anhydrase, SULFATE ION, ... | | Authors: | Marapaka, A.K, Das, C, Flaherty, D.P, Yadav, R. | | Deposit date: | 2022-07-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Characterization of Thiadiazolesulfonamide Inhibitors Bound to Neisseria gonorrhoeae alpha-Carbonic Anhydrase.
Acs Med.Chem.Lett., 14, 2023
|
|
8BBP
 
 | |
6YF1
 
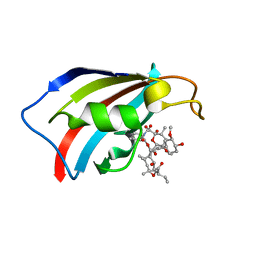 | | FKBP12 in complex with the BMP potentiator compound 8 at 1.12A resolution | | Descriptor: | (1aR,3R,5S,6R,7S,9R,10R,17aS,20S,21R,22S,25R,25aR)-25-Ethyl-10,22-dihydroxy-20-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-5,7-dimethoxy-1a,3,9,21-tetramethyloctadecahydro-2H-6,10-epoxyoxireno[p]pyrido[2,1-c][1,4]oxazacyclotricosine-11,12,18,24(1aH,14H)-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6X25
 
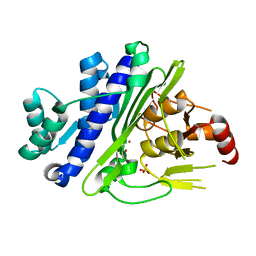 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AND LITHIUM AT 3.2 ANGSTROM RESOLUTION | | Descriptor: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Dollins, D.E, Endo-Streeter, S, Ren, Y, York, J.D. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
5KH7
 
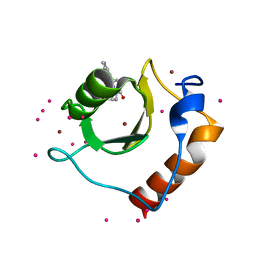 | | Crystal structure of fragment (3-[6-Oxo-3-(3-pyridinyl)-1(6H)-pyridazinyl]propanoic acid) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(6-oxidanylidene-3-pyridin-3-yl-pyridazin-1-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Walker, J, Ravichandran, M, Ferreira de Freitas, R, Schapira, M, Bountra, C, Edwards, A.M, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
6UAB
 
 | |
8P86
 
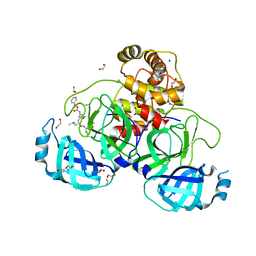 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 mM MG-132, from an "old" crystal. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
5GYB
 
 | |
5KGY
 
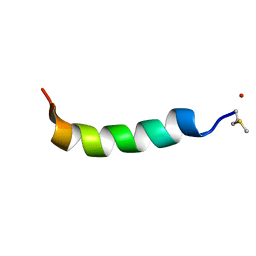 | | Phenol-soluble modulin Alpha 3 | | Descriptor: | Phenol-soluble modulin alpha 3 peptide | | Authors: | Towle, K.M, Lohans, C.T, Acedo, J.Z, Van Belkum, M.J, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Phenol-Soluble Modulins alpha 1, alpha 3, and beta 2, Virulence Factors from Staphylococcus aureus.
Biochemistry, 55, 2016
|
|
5GYE
 
 | |
7V5N
 
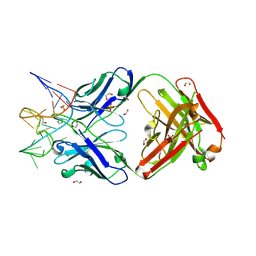 | | Crystal structure of Fab fragment of bevacizumab bound to DNA aptamer | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*GP*TP*TP*GP*GP*TP*GP*GP*TP*AP*GP*TP*TP*AP*CP*GP*TP*TP*CP*GP*C)-3'), IMIDAZOLE, ... | | Authors: | Hishiki, A, Tong, J, Todoroki, K, Hashimoto, H. | | Deposit date: | 2021-08-17 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a DNA aptamer that binds to the complementarity-determining region of therapeutic monoclonal antibody and affinity improvement induced by pH-change for sensitive detection.
Biosens.Bioelectron., 203, 2022
|
|
