4HW1
 
 | | Multiple Crystal structures of an all-AT DNA dodecamer stabilized by weak interactions. | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*TP*TP*TP*AP*TP*T)-3'), MAGNESIUM ION | | Authors: | Acosta-Reyes, F, Subirana, J.A, Pous, J, Condom, N, Malinina, L, Campos, J.L. | | Deposit date: | 2012-11-07 | | Release date: | 2013-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Polymorphic crystal structures of an all-AT DNA dodecamer.
Biopolymers, 103, 2015
|
|
4X9J
 
 | | EGR-1 with Doubly Methylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*(5CM)P*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*(5CM)P*GP*C)-3'), Early growth response protein 1, ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2014-12-11 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1.
Febs Lett., 589, 2015
|
|
2ELG
 
 | | The rare crystallographic structure of d(CGCGCG)2: The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)2 at 10 degree celsius | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DG)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ohishi, H, Tozuka, Y, Zhou, D.Y, Ishida, T, Nakatani, K. | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The rare crystallographic structure of d(CGCGCG)(2): The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)(2) at 10 degrees C
Biochem.Biophys.Res.Commun., 358, 2007
|
|
8V32
 
 | | TnsD-TnsC-DNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (41-MER), MAGNESIUM ION, ... | | Authors: | Chang, L, Wang, S. | | Deposit date: | 2023-11-26 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of TnsABCD transpososome reveals mechanisms of targeted DNA transposition.
Cell, 2024
|
|
115D
 
 | | ORDERED WATER STRUCTURE IN AN A-DNA OCTAMER AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*GP*(BRU)P*AP*(BRU)P*AP*CP*C)-3') | | Authors: | Kennard, O, Cruse, W.B.T, Nachman, J, Prange, T, Shakked, Z, Rabinovich, D. | | Deposit date: | 1993-02-12 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ordered water structure in an A-DNA octamer at 1.7 A resolution.
J.Biomol.Struct.Dyn., 3, 1986
|
|
5CDR
 
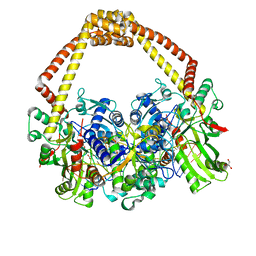 | | 2.65 structure of S.aureus DNA gyrase and artificially nicked DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*)-3'), DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
3P57
 
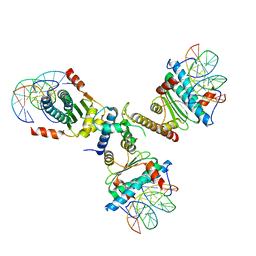 | | Crystal structure of the p300 TAZ2 domain bound to MEF2 on DNA | | Descriptor: | DNA (5'-D(*A*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), Histone acetyltransferase p300, ... | | Authors: | He, J, Ye, J, Riquelme, C, Liu, J.O. | | Deposit date: | 2010-10-08 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1921 Å) | | Cite: | Structure of p300 bound to MEF2 on DNA reveals a mechanism of enhanceosome assembly.
Nucleic Acids Res., 39, 2011
|
|
8HNI
 
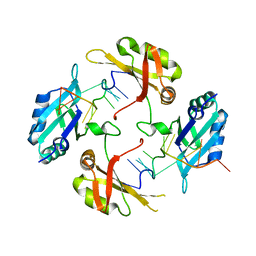 | | hnRNP A2/B1 RRMs in complex with telomeric DNA | | Descriptor: | DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Liu, Y, Abula, A, Xiao, H, Guo, H, Li, T, Zheng, L, Chen, B, Nguyen, H, Ji, X. | | Deposit date: | 2022-12-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2023
|
|
3VW3
 
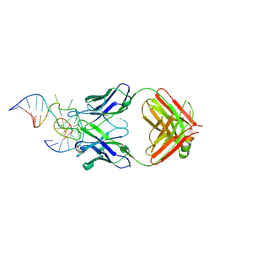 | | Antibody 64M-5 Fab in complex with a double-stranded DNA (6-4) photoproduct | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain), COBALT HEXAMMINE(III), ... | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y. | | Deposit date: | 2012-07-30 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a double-stranded DNA (6-4) photoproduct in complex with the 64M-5 antibody Fab
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5X6G
 
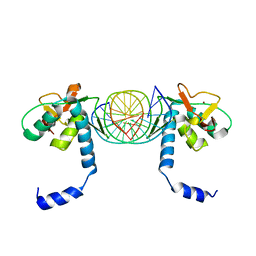 | | Crystal Structure of SMAD5-MH1/palindromic SBE DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*GP*TP*AP*TP*GP*TP*CP*TP*AP*GP*AP*CP*TP*GP*A)-3'), Mothers against decapentaplegic homolog 5, ... | | Authors: | Chai, N, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for the Smad5 MH1 domain to recognize different DNA sequences.
Nucleic Acids Res., 43, 2015
|
|
2G7H
 
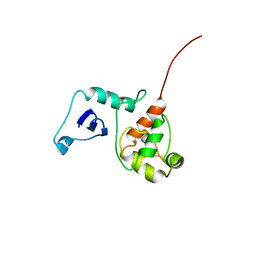 | |
1XWL
 
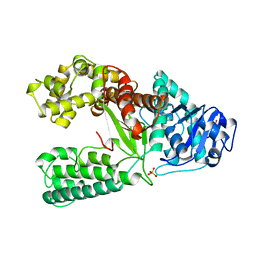 | |
1C35
 
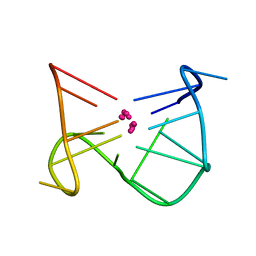 | | SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMIDIATE | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Bolton, P.H, Marathias, V.M, Wang, K. | | Deposit date: | 1999-07-25 | | Release date: | 1999-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA.
Nucleic Acids Res., 28, 2000
|
|
1OMH
 
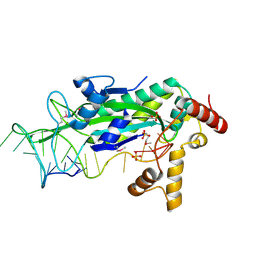 | | Conjugative Relaxase TrwC in complex with OriT Dna. Metal-free structure. | | Descriptor: | DNA OLIGONUCLEOTIDE, SULFATE ION, trwC protein | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2003-02-25 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC.
Nat.Struct.Biol., 10, 2003
|
|
282D
 
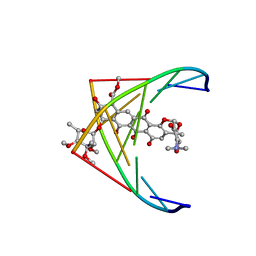 | | A CONTINOUS TRANSITION FROM A-DNA TO B-DNA IN THE 1:1 COMPLEX BETWEEN NOGALAMYCIN AND THE HEXAMER DCCCGGG | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*G)-3'), NOGALAMYCIN | | Authors: | Cruse, W, Saludjian, P, Leroux, Y, Leger, Y, El Manouni, D, Prange, T. | | Deposit date: | 1996-08-26 | | Release date: | 1996-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A continuous transition from A-DNA to B-DNA in the 1:1 complex between nogalamycin and the hexamer dCCCGGG.
J.Biol.Chem., 271, 1996
|
|
382D
 
 | |
1D49
 
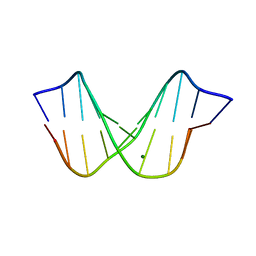 | | THE STRUCTURE OF A B-DNA DECAMER WITH A CENTRAL T-A STEP: C-G-A-T-T-A-A-T-C-G | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*AP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Grzeskowiak, K, Yanagi, K, Dickerson, R.E. | | Deposit date: | 1991-09-17 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G.
J.Mol.Biol., 225, 1992
|
|
6RYI
 
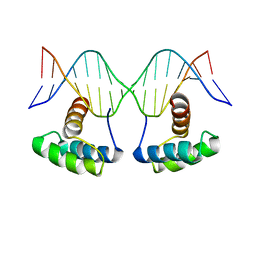 | | WUS-HD bound to G-Box DNA | | Descriptor: | DNA (5'-D(P*CP*CP*CP*AP*TP*CP*AP*CP*GP*TP*GP*AP*CP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*TP*CP*AP*CP*GP*TP*GP*AP*TP*GP*GP*G)-3'), Protein WUSCHEL | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
213D
 
 | |
209D
 
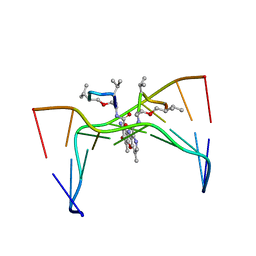 | | Structural, physical and biological characteristics of RNA:DNA binding agent N8-actinomycin D | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3'), N8-ACTINOMYCIN D | | Authors: | Shinomiya, M, Chu, W, Carlson, R.G, Weaver, R.F, Takusagawa, F. | | Deposit date: | 1995-05-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural, Physical, and Biological Characteristics of RNA.DNA Binding Agent N8-Actinomycin D.
Biochemistry, 34, 1995
|
|
279D
 
 | |
2LEF
 
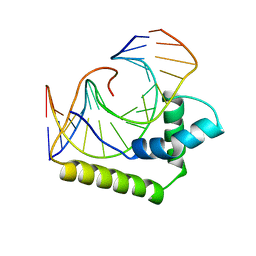 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | Authors: | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
142D
 
 | |
141D
 
 | |
117D
 
 | |
