3MIA
 
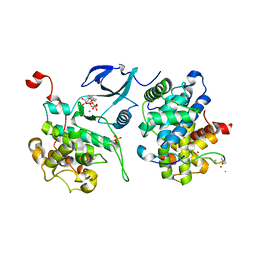 | | Crystal structure of HIV-1 Tat complexed with ATP-bound human P-TEFb | | Descriptor: | Cell division protein kinase 9, Cyclin-T1, MAGNESIUM ION, ... | | Authors: | Tahirov, T.H, Babayeva, N.D, Varzavand, K, Cooper, J.J, Sedore, S.C, Price, D.H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb.
Nature, 465, 2010
|
|
4OR5
 
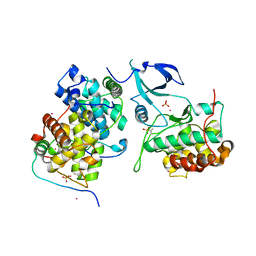 | | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4 | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Gu, J, Babayeva, N.D, Suwa, Y, Baranovskiy, A.G, Price, D.H, Tahirov, T.H. | | Deposit date: | 2014-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4.
Cell Cycle, 13, 2014
|
|
3MY1
 
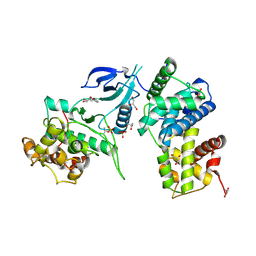 | | Structure of CDK9/cyclinT1 in complex with DRB | | Descriptor: | 5,6-dichloro-1-beta-D-ribofuranosyl-1H-benzimidazole, Cell division protein kinase 9, Cyclin-T1, ... | | Authors: | Baumli, S, Johnson, L.N. | | Deposit date: | 2010-05-09 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Halogen bonds form the basis for selective P-TEFb inhibition by DRB
Chem.Biol., 17, 2010
|
|
3MI9
 
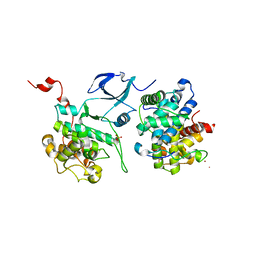 | | Crystal structure of HIV-1 Tat complexed with human P-TEFb | | Descriptor: | Cell division protein kinase 9, Cyclin-T1, Protein Tat, ... | | Authors: | Tahirov, T.H, Babayeva, N.D, Varzavand, K, Cooper, J.J, Sedore, S.C, Price, D.H. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb.
Nature, 465, 2010
|
|
4OGR
 
 | |
3D2M
 
 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-glutamate | | Descriptor: | COENZYME A, GLUTAMIC ACID, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
3D2P
 
 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-arginine | | Descriptor: | ARGININE, COENZYME A, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
4ZUX
 
 | |
7B3A
 
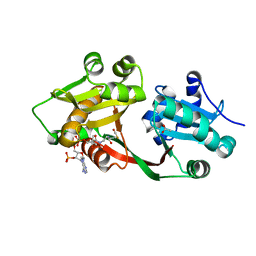 | | Crystal structure of PamZ | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Loll, B, Dang, T, Mainz, A, Suessmuth, R, Wahl, M.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Molecular basis of antibiotic self-resistance in a bee larvae pathogen.
Nat Commun, 13, 2022
|
|
4IMY
 
 | |
4H6U
 
 | | Tubulin acetyltransferase mutant | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase, PHOSPHATE ION, ... | | Authors: | Roll-Mecak, A, Kizub, V, Szyk, A. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4482 Å) | | Cite: | Crystal structures of tubulin acetyltransferase reveal a conserved catalytic core and the plasticity of the essential N terminus.
J.Biol.Chem., 287, 2012
|
|
4H6Z
 
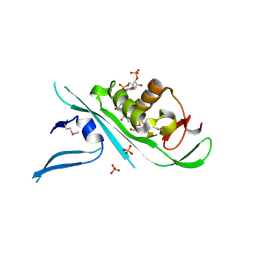 | | Tubulin acetyltransferase | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase, PHOSPHATE ION | | Authors: | Kizub, L, Szyk, A, Piszczek, G, Roll-Mecak, A. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of tubulin acetyltransferase reveal a conserved catalytic core and the plasticity of the essential N terminus.
J.Biol.Chem., 287, 2012
|
|
5XUN
 
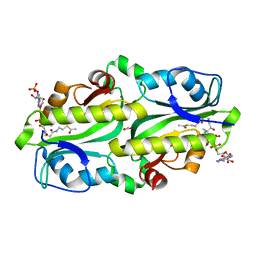 | | Crystal structure of Y145F mutant of KacT | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, Acetyltransferase | | Authors: | Qian, H.L, Yao, Q.Q, Gan, J.H, Ou, H.Y. | | Deposit date: | 2017-06-23 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and characterization of acetyltransferase-type toxin-antitoxin locus in Klebsiella pneumoniae
Mol. Microbiol., 108, 2018
|
|
6MFD
 
 | | GphF GNAT-like decarboxylase in complex with isobutyryl-CoA | | Descriptor: | ACETATE ION, GLYCEROL, GphF, ... | | Authors: | Skiba, M.A, Tran, C.L, Smith, J.L. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Repurposing the GNAT Fold in the Initiation of Polyketide Biosynthesis.
Structure, 28, 2020
|
|
6MFC
 
 | | GphF GNAT-like decarboxylase | | Descriptor: | GLYCEROL, GphF, PENTAETHYLENE GLYCOL | | Authors: | Skiba, M.A, Tran, C.L, Smith, J.L. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.589 Å) | | Cite: | Repurposing the GNAT Fold in the Initiation of Polyketide Biosynthesis.
Structure, 28, 2020
|
|
9G7F
 
 | |
9G79
 
 | |
7K09
 
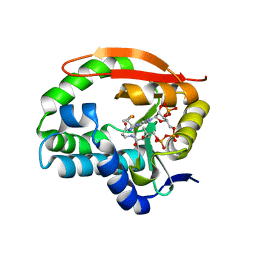 | | Puromycin N-acetyltransferase in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Puromycin N-acetyltransferase | | Authors: | Caputo, A.T, Newman, J, Adams, T.E, Peat, T.S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins.
Sci Rep, 11, 2021
|
|
7K0A
 
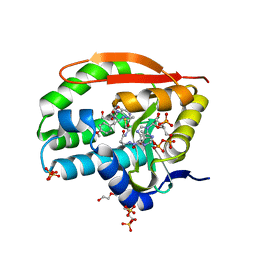 | | Puromycin N-acetyltransferase in complex with acetylated puromycin and CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Caputo, A.T, Newman, J, Adams, T.E. | | Deposit date: | 2020-09-03 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins.
Sci Rep, 11, 2021
|
|
1XMT
 
 | | X-ray structure of gene product from arabidopsis thaliana at1g77540 | | Descriptor: | BROMIDE ION, putative acetyltransferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
7KWQ
 
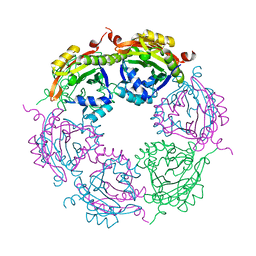 | | Spermidine N-acetyltransferase SpeG R149-K152 chimera from Vibrio cholerae and hSSAT | | Descriptor: | Spermidine N(1)-acetyltransferase | | Authors: | Le, V.T.B, Tsimbalyuk, S, Lim, E.Q, Solis, A, Gawat, D, Boeck, P, Renolo, R, Forwood, J.K, Kuhn, M.L. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Vibrio cholerae SpeG Spermidine/Spermine N -Acetyltransferase Allosteric Loop and beta 6-beta 7 Structural Elements Are Critical for Kinetic Activity.
Front Mol Biosci, 8, 2021
|
|
6T9J
 
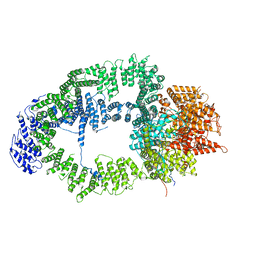 | | SAGA Tra1 module | | Descriptor: | Transcription factor SPT20, Transcription initiation factor TFIID subunit 12, Transcription-associated protein 1 | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6T9I
 
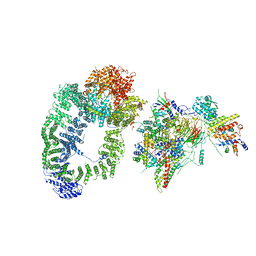 | | cryo-EM structure of transcription coactivator SAGA | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6IG9
 
 | |
6T9L
 
 | |
