4V99
 
 | |
4V8H
 
 | | Crystal structure of HPF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Hibernation Factors RMF, HPF, and YfiA Turn Off Protein Synthesis.
Science, 336, 2012
|
|
4V9F
 
 | |
4UXL
 
 | | Structure of Human ROS1 Kinase Domain in Complex with PF-06463922 | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ROS | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2014-08-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pf-06463922 is a Potent and Selective Next-Generation Ros1/Alk Inhibitor Capable of Blocking Crizotinib-Resistant Ros1 Mutations.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4V67
 
 | | Crystal structure of a translation termination complex formed with release factor RF2. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Asahara, H, Lancaster, L, Laurberg, M, Hirschi, A, Noller, H.F. | | Deposit date: | 2008-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a translation termination complex formed with release factor RF2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4V8D
 
 | | Structure analysis of ribosomal decoding (cognate tRNA-tyr complex). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V95
 
 | | Crystal structure of YAEJ bound to the 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Gagnon, M.G, Seetharaman, S.V, Bulkley, D.P, Steitz, T.A. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome.
Science, 335, 2012
|
|
4V9S
 
 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
6UFP
 
 | | Structure of proline utilization A with the FAD covalently modified by L-thiazolidine-2-carboxylate and three cysteines (Cys46, Cys470, Cys638) modified to S,S-(2-HYDROXYETHYL)THIOCYSTEINE | | Descriptor: | (2S)-1,3-thiazolidine-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Campbell, A.C, Tanner, J.J. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Covalent Modification of the Flavin in Proline Dehydrogenase by Thiazolidine-2-Carboxylate.
Acs Chem.Biol., 15, 2020
|
|
4V51
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with mRNA, tRNA and paromomycin | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Selmer, M, Dunham, C.M, Murphy, F.V, Weixlbaumer, A, Petry, S, Weir, J.R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2006-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70S ribosome complexed with mRNA and tRNA.
Science, 313, 2006
|
|
7PK5
 
 | |
7QWR
 
 | | Structure of the ribosome-nascent chain containing an ER signal sequence in complex with NAC | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
7QWQ
 
 | | Ternary complex of ribosome nascent chain with SRP and NAC | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Gamerdinger, M, Hsieh, H, Wallisch, A, Chandrasekaran, V, Ulusoy, Z, Scaiola, A, Hegde, R, Shan, S, Ban, N, Deuerling, E. | | Deposit date: | 2022-01-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting.
Science, 375, 2022
|
|
7QA0
 
 | |
7QA3
 
 | |
7QAV
 
 | |
4UG0
 
 | | STRUCTURE OF THE HUMAN 80S RIBOSOME | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S RIBOSOMAL PROTEIN, ... | | Authors: | Khatter, H, Myasnikov, A.G, Natchiar, S.K, Klaholz, B.P. | | Deposit date: | 2015-03-20 | | Release date: | 2015-06-10 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the human 80S ribosome
NATURE, 520, 2015
|
|
7PB4
 
 | | Cenp-HIK 3-protein complex | | Descriptor: | Centromere protein H, Centromere protein I, Centromere protein K | | Authors: | Bellini, D, Yatskevich, S, Muir, W.K, Barford, D. | | Deposit date: | 2021-07-30 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
6KO7
 
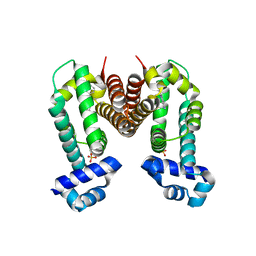 | | Crystal structure of the Ethidium bound RamR determined with XtaLAB Synergy | | Descriptor: | ETHIDIUM, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
5M25
 
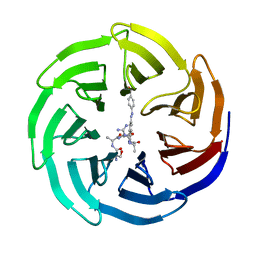 | | Modulation of MLL1 Methyltransferase Activity | | Descriptor: | (2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-azanyl-3-oxidanyl-propanoyl]amino]propanoyl]amino]-5-carbamimidamido-~{N}-[(2~{S})-1-[[4-[(~{E})-[4-(hydroxymethyl)phenyl]diazenyl]phenyl]methylamino]-1-oxidanylidene-propan-2-yl]pentanamide, WD repeat-containing protein 5 | | Authors: | Srinivasan, V. | | Deposit date: | 2016-10-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Controlled inhibition of methyltransferases using photoswitchable peptidomimetics: towards an epigenetic regulation of leukemia.
Chem Sci, 8, 2017
|
|
3L3M
 
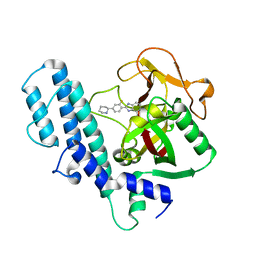 | | PARP complexed with A927929 | | Descriptor: | 2-{2-fluoro-4-[(2S)-piperidin-2-yl]phenyl}-1H-benzimidazole-7-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-12-17 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization of phenyl-substituted benzimidazole carboxamide poly(ADP-ribose) polymerase inhibitors: identification of (S)-2-(2-fluoro-4-(pyrrolidin-2-yl)phenyl)-1H-benzimidazole-4-carboxamide (A-966492), a highly potent and efficacious inhibitor.
J.Med.Chem., 53, 2010
|
|
6KO9
 
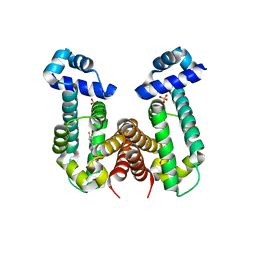 | | Crystal structure of the Gefitinib Intermediate 1 bound RamR determined with XtaLAB Synergy | | Descriptor: | 4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-methoxy-quinazolin-6-ol, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
2QPY
 
 | | AR LBD with small molecule | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Coactivator peptide, ... | | Authors: | Estebanez-Perpina, E, Fletterick, R. | | Deposit date: | 2007-07-25 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6KO8
 
 | | Crystal structure of the Cholic acid bound RamR determined with XtaLAB Synergy | | Descriptor: | CHOLIC ACID, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
3L3L
 
 | | PARP complexed with A906894 | | Descriptor: | 3-oxo-2-piperidin-4-yl-2,3-dihydro-1H-isoindole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | Authors: | Park, C.H. | | Deposit date: | 2009-12-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and SAR of substituted 3-oxoisoindoline-4-carboxamides as potent inhibitors of poly(ADP-ribose) polymerase (PARP) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
