7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
6MP6
 
 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
5FXJ
 
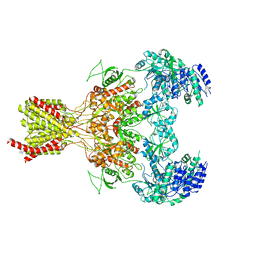 | | GluN1b-GluN2B NMDA receptor structure-Class X | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, NMDA 1, NMDA 2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa H, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5LQX
 
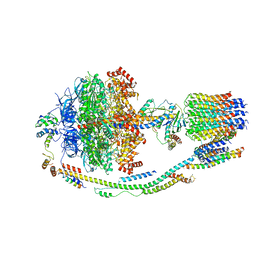 | | Structure of F-ATPase from Pichia angusta, state3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase AAP1 subunit, ... | | Authors: | Vinothkumar, K.R, Montgomery, M.G, Liu, S, Walker, J.E. | | Deposit date: | 2016-08-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mitochondrial ATP synthase fromPichia angustadetermined by electron cryo-microscopy.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
8AT5
 
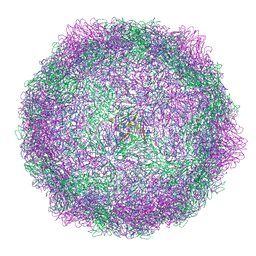 | | native Coxsackievirus A9 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Domanska, A, Plavec, Z, Ruokolainen, V, Marjomaki, V.S, Butcher, S.J. | | Deposit date: | 2022-08-22 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Studies Reveal that Endosomal Cations Promote Formation of Infectious Coxsackievirus A9 A-Particles, Facilitating RNA and VP4 Release.
J.Virol., 96, 2022
|
|
1RPN
 
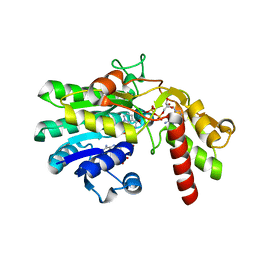 | | Crystal Structure of GDP-D-mannose 4,6-dehydratase in complexes with GDP and NADPH | | Descriptor: | GDP-mannose 4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Webb, N.A, Mulichak, A.M, Lam, J.S, Rocchetta, H.L, Garavito, R.M. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a tetrameric GDP-D-mannose 4,6-dehydratase from a bacterial GDP-D-rhamnose biosynthetic pathway.
Protein Sci., 13, 2004
|
|
5FFR
 
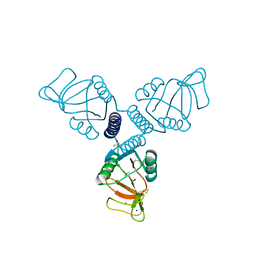 | | Crystal Structure of Surfactant Protein-A complexed with phosphocholine | | Descriptor: | CALCIUM ION, PHOSPHOCHOLINE, Pulmonary surfactant-associated protein A, ... | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
6HLU
 
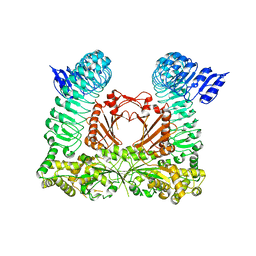 | |
8ADQ
 
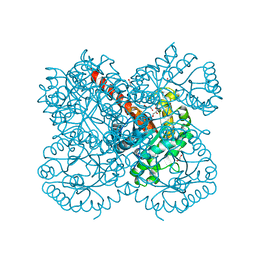 | | Crystal structure of holo-SwHPA-Mg (hydroxy ketone aldolase) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate and D-Glyceraldehyde | | Descriptor: | 3-HYDROXYPYRUVIC ACID, BROMIDE ION, D-Glyceraldehyde, ... | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8T6I
 
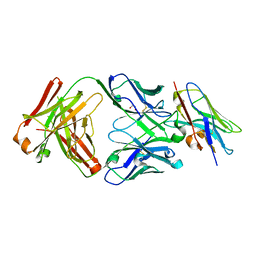 | | Structure of VHH-Fab complex with engineered Crystal Kappa region | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Filippova, E.V, Thompson, I, Kossiakoff, A.A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
6HMT
 
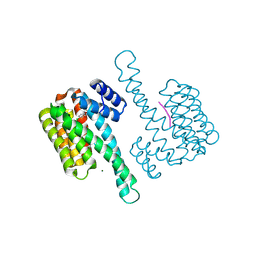 | | Ternary complex of Estrogen Receptor alpha peptide and 14-3-3 sigma C42 mutant bound to disulfide fragment PPI stabilizer 2 | | Descriptor: | 14-3-3 protein sigma, 2-(4-chloranylphenoxy)-2-methyl-~{N}-(2-sulfanylethyl)propanamide, Estrogen Receptor, ... | | Authors: | Sijbesma, E, Hallenbeck, K.K, Leysen, S, Arkin, M.R, Ottmann, C. | | Deposit date: | 2018-09-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Site-Directed Fragment-Based Screening for the Discovery of Protein-Protein Interaction Stabilizers.
J. Am. Chem. Soc., 141, 2019
|
|
6SVH
 
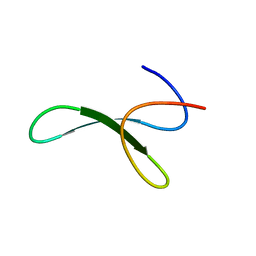 | | Protein allostery of the WW domain at atomic resolution: FFpSPR bound structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8TS5
 
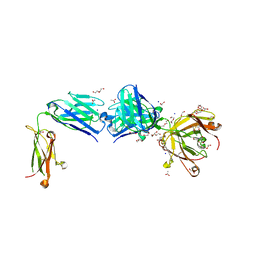 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
1C3D
 
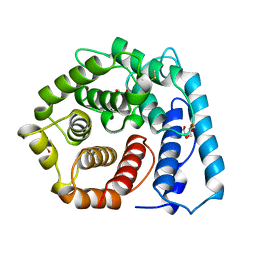 | | X-RAY CRYSTAL STRUCTURE OF C3D: A C3 FRAGMENT AND LIGAND FOR COMPLEMENT RECEPTOR 2 | | Descriptor: | C3D, GLYCEROL | | Authors: | Nagar, B, Jones, R.G, Diefenbach, R.J, Isenman, D.E, Rini, J.M. | | Deposit date: | 1998-05-19 | | Release date: | 1998-10-07 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2.
Science, 280, 1998
|
|
8IF8
 
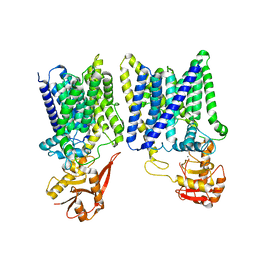 | | Arabinosyltransferase AftA | | Descriptor: | CALCIUM ION, Galactan 5-O-arabinofuranosyltransferase | | Authors: | Gong, Y.C, Rao, Z.H, Zhang, L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the priming arabinosyltransferase AftA required for AG biosynthesis of Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SXS
 
 | |
5ITW
 
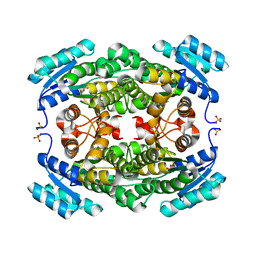 | | Crystal structure of Bacillus subtilis BacC Dihydroanticapsin 7-dehydrogenase | | Descriptor: | Dihydroanticapsin 7-dehydrogenase, SULFATE ION | | Authors: | Perinbam, K, Balaram, H, Row, T.N.G, Gopal, B. | | Deposit date: | 2016-03-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Probing the influence of non-covalent contact networks identified by charge density analysis on the oxidoreductase BacC.
Protein Eng. Des. Sel., 30, 2017
|
|
8T7I
 
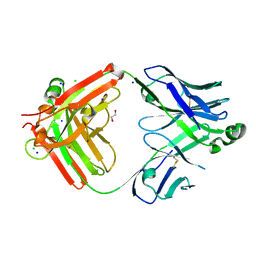 | | Structure of the S1CE variant of Fab F1 (FabS1CE-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7G
 
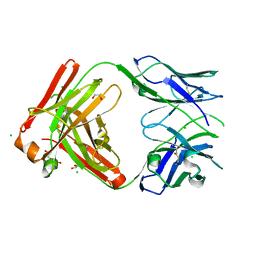 | | Structure of the CK variant of Fab F1 (FabC-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CK variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRT
 
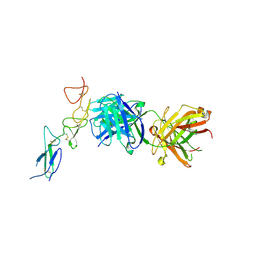 | | Structure of the EphA2 CRD bound to FabS1CE_C1, monoclinic form | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 2, S1CE variant of Fab C1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
5ITV
 
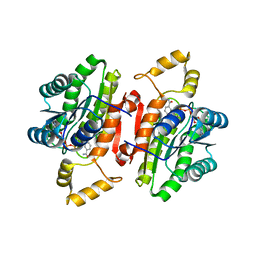 | | Crystal structure of Bacillus subtilis BacC Dihydroanticapsin 7-dehydrogenase in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dihydroanticapsin 7-dehydrogenase | | Authors: | Perinbam, K, Balaram, H, Row, T.N.G, Gopal, B. | | Deposit date: | 2016-03-17 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing the influence of non-covalent contact networks identified by charge density analysis on the oxidoreductase BacC.
Protein Eng. Des. Sel., 30, 2017
|
|
6HCH
 
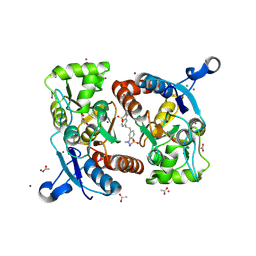 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-L504Y-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM01 AT 1.6 A RESOLUTION. | | Descriptor: | 6,6'-(Ethane-1,2-diyl)bis(4-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide), ACETATE ION, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-15 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
8TRS
 
 | | Structure of the EphA2 CRD bound to FabS1CE_C1, trigonal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7F
 
 | | Structure of the S1 variant of Fab F1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S1 variant of Fab F1 heavy chain, S1 variant of Fab F1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
