5TWO
 
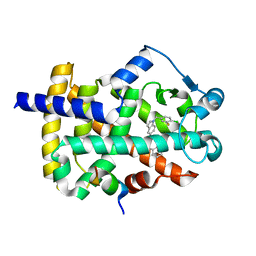 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selectively PPAR gamma-modulating ligand VSP-51 | | Descriptor: | N-benzyl-1-[(4-chloro-3-fluorophenyl)methyl]-1H-indole-5-carboxamide, PRO-SER-LEU-LEU-LYS-LYS-LEU-LEU-LEU-ALA-PRO, Peroxisome proliferator-activated receptor gamma | | Authors: | Yi, W, Shi, J, Zhao, G, Zhou, X.E, Suino-Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2016-11-14 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Identification of a novel selective PPAR gamma ligand with a unique binding mode and improved therapeutic profile in vitro.
Sci Rep, 7, 2017
|
|
8DUG
 
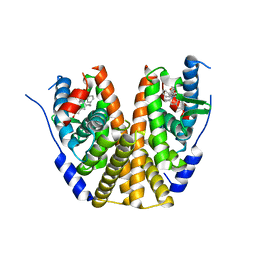 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with (6'-hydroxy-1'-(4-(2-((S)-3-methylpyrrolidin-1-yl)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-(4-{2-[(3R)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
8DUB
 
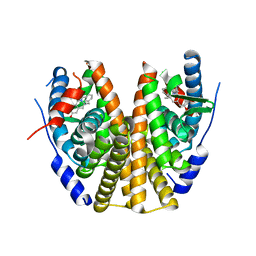 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with ((1'-(4-((1-ethylpyrrolidin-3-yl)methyl)phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-1'-(4-{[(3S)-1-ethylpyrrolidin-3-yl]oxy}phenyl)-6'-hydroxy-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Hancock, G.R, Young, K.S, Hosfield, D.J, Joiner, C, Sullivan, E.A, Yildz, Y, Laine, M, Greene, G.L, Fanning, S.W. | | Deposit date: | 2022-07-27 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells.
NPJ Breast Cancer, 8, 2022
|
|
7THZ
 
 | |
7THY
 
 | |
1U3H
 
 | | Crystal structure of mouse TCR 172.10 complexed with MHC class II I-Au molecule at 2.4 A | | Descriptor: | H-2 class II histocompatibility antigen, A-U alpha chain, A-U beta chain, ... | | Authors: | Maynard, J, Petersson, K, Wilson, D.H, Adams, E.J, Blondelle, S.E, Boulanger, M.J, Wilson, D.B, Garcia, K.C. | | Deposit date: | 2004-07-21 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity
Immunity, 22, 2005
|
|
8EAQ
 
 | | Structure of the full-length IP3R1 channel determined at high Ca2+ | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 1, ... | | Authors: | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
8EAR
 
 | | Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
6JKQ
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi (Ligand-free form) | | Descriptor: | Aspartate carbamoyltransferase | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6VZG
 
 | | Cryo-EM structure of Sth1-Arp7-Arp9-Rtt102 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-like protein ARP9, Actin-related protein 7, ... | | Authors: | Leschziner, A.E, Baker, R.W. | | Deposit date: | 2020-02-28 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into assembly and function of the RSC chromatin remodeling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6VPB
 
 | |
5OC9
 
 | | Crystal Structure of human TMEM16K / Anoctamin 10 | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
4ZU7
 
 | | X-ray structure if the QdtA 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum, double mutant Y17R/R97H, in complex with TDP | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, QdtA, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Thoden, J.B, Vinogradov, E, Gilbert, M, Salinger, A.J, Holden, H.M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial Sugar 3,4-Ketoisomerases: Structural Insight into Product Stereochemistry.
Biochemistry, 54, 2015
|
|
3SUA
 
 | | Crystal structure of the intracellular domain of Plexin-B1 in complex with Rac1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-B1, ... | | Authors: | Bell, C.H, Aricescu, A.R, Jones, E.Y, Siebold, C. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.39 Å) | | Cite: | A Dual Binding Mode for RhoGTPases in Plexin Signalling.
Plos Biol., 9, 2011
|
|
4I0C
 
 | | The structure of the camelid antibody cAbHuL5 in complex with human lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | De Genst, E, Chan, P.H, Pardon, E, Kumita, J.R, Christodoulou, J, Menzer, L, Chirgadze, D.Y, Robinson, C.V, Muyldermans, S, Matagne, A, Wyns, L, Dobson, C.M, Dumoulin, M. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-09 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nanobody binding to non-amyloidogenic regions of the protein human lysozyme enhances partial unfolding but inhibits amyloid fibril formation.
J.Phys.Chem.B, 117, 2013
|
|
8DYG
 
 | | IL17A homodimer bound to Compound 7 | | Descriptor: | (5P)-2-hydroxy-5-(6-methylquinolin-5-yl)benzoic acid, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
2JEL
 
 | | JEL42 FAB/HPR COMPLEX | | Descriptor: | HISTIDINE-CONTAINING PROTEIN, JEL42 FAB FRAGMENT, SULFATE ION | | Authors: | Prasad, L, Waygood, E.B, Lee, J.S, Delbaere, L.T.J. | | Deposit date: | 1998-02-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A resolution structure of the jel42 Fab fragment/HPr complex
J.Mol.Biol., 280, 1998
|
|
5A3Z
 
 | | Structure of monoclinic Lysozyme obtained by multi crystal data collection | | Descriptor: | GLYCEROL, LYSOZYME, NITRATE ION | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-04 | | Release date: | 2015-11-11 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8EAD
 
 | |
2W6V
 
 | | Structure of Human deoxy Hemoglobin A in complex with Xenon | | Descriptor: | GLYCEROL, HEMOGLOBIN SUBUNIT ALPHA, HEMOGLOBIN SUBUNIT BETA, ... | | Authors: | Miele, A.E, Draghi, F, Sciara, G, Johnson, K.A, Renzi, F, Vallone, B, Brunori, M, Savino, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pattern of Cavities in Globins: The Case of Human Hemoglobin.
Biopolymers, 91, 2009
|
|
5S4O
 
 | | Tubulin-Z48847594-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muehlethaler, T, Gioia, D, Prota, A.E, Sharpe, M.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2020-11-08 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comprehensive Analysis of Binding Sites in Tubulin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2C8E
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (Free state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
3ZRD
 
 | | Oxidised Thiol peroxidase (Tpx) from Yersinia pseudotuberculosis | | Descriptor: | THIOL PEROXIDASE | | Authors: | Gabrielsen, M, Zetterstrom, C.E, Wang, D, Elofsson, M, Roe, A.J. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Characterisation of Tpx from Yersinia Pseudotuberculosis Reveals Insights Into the Binding of Salicylidene Acylhydrazide Compounds.
Plos One, 7, 2012
|
|
5S4Q
 
 | | Tubulin-Z422344882-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(4-methyl-1,3-thiazole-5-carbonyl)piperazin-2-one, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Gioia, D, Prota, A.E, Sharpe, M.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2020-11-08 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comprehensive Analysis of Binding Sites in Tubulin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7KPY
 
 | | Crystal structure of CBP bromodomain liganded with UMB298 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloranyl-4-methoxy-phenyl)ethyl]-~{N}-cyclohexyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)imidazo[1,2-a]pyridin-3-amine, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Dimethylisoxazole-Attached Imidazo[1,2- a ]pyridines as Potent and Selective CBP/P300 Inhibitors.
J.Med.Chem., 64, 2021
|
|
