5SK3
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with [2-cyclopropyl-6-[2-(1-methyl-4-phenylimidazol-2-yl)ethyl]imidazo[1,2-b]pyridazin-3-yl]methanol | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SE9
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 2,3-dimethyl-6-[(1-methyl-4-phenylimidazol-2-yl)methoxy]imidazo[1,2-b]pyridazine | | Descriptor: | (4R)-2,3-dimethyl-6-[(1-methyl-4-phenyl-1H-imidazol-2-yl)methoxy]imidazo[1,2-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
1SGR
 
 | |
5SGN
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 2,3-dimethyl-6-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-7-(trifluoromethyl)imidazo[1,2-b]pyridazine | | Descriptor: | (4S)-2,3-dimethyl-6-{2-[1-methyl-3-(pyrrolidin-1-yl)-1H-1,2,4-triazol-5-yl]ethyl}-7-(trifluoromethyl)imidazo[1,2-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SH3
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 2,3-dimethyl-6-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-8-methylsulfonylimidazo[1,2-b]pyridazine | | Descriptor: | (4S)-8-(methanesulfonyl)-2,3-dimethyl-6-{2-[1-methyl-3-(pyrrolidin-1-yl)-1H-1,2,4-triazol-5-yl]ethyl}imidazo[1,2-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SIB
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 3-methyl-6-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-2-(trifluoromethyl)imidazo[1,2-b]pyridazine | | Descriptor: | (4R)-3-methyl-6-{2-[1-methyl-3-(pyrrolidin-1-yl)-1H-1,2,4-triazol-5-yl]ethyl}-2-(trifluoromethyl)imidazo[1,2-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
3EIU
 
 | | A second transient position of ATP on its trail to the nucleotide-binding site of subunit B of the motor protein A1Ao ATP synthase | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ADENOSINE-5'-TRIPHOSPHATE, V-type ATP synthase beta chain | | Authors: | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2008-09-17 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | A second transient position of ATP on its trail to the nucleotide-binding site of subunit B of the motor protein A(1)A(O) ATP synthase
J.Struct.Biol., 166, 2009
|
|
2SGP
 
 | | PRO 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | OVOMUCOID INHIBITOR, PHOSPHATE ION, PROTEINASE B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
4OR6
 
 | |
5BV6
 
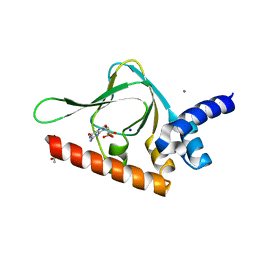 | | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with cGMP | | Descriptor: | ACETATE ION, CALCIUM ION, GUANOSINE-3',5'-MONOPHOSPHATE, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-04 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
1T3R
 
 | | HIV protease wild-type in complex with TMC114 inhibitor | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, PHOSPHATE ION, protease RETROPEPSIN | | Authors: | King, N.M, Prabu-Jeyabalan, M, Nalivaika, E.A, Wigernick, P.B, de Bethune, M.P, Schiffer, C.A. | | Deposit date: | 2004-04-27 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor
J.Med.Chem., 48, 2005
|
|
1B9T
 
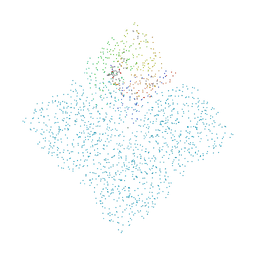 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 1-(4-CARBOXY-2-GUANIDINOPENTYL)-5,5'-DI(HYDROXYMETHYL)PYRROLIDIN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
7KAA
 
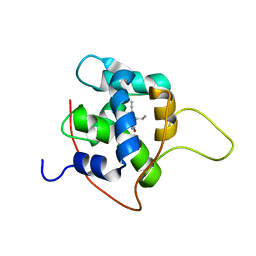 | | NMR solution structures of tirasemtiv drug bound to a fast skeletal troponin C-troponin I complex | | Descriptor: | 6-ethynyl-1-(pentan-3-yl)-1H-imidazo[4,5-b]pyrazin-2-ol, CALCIUM ION, Troponin C, ... | | Authors: | Mercier, P, Li, M.X, Hartman, J.J, Sykes, B.D. | | Deposit date: | 2020-09-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Tirasemtiv Activation of Fast Skeletal Muscle.
J.Med.Chem., 64, 2021
|
|
3W11
 
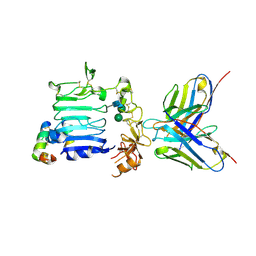 | |
1T7I
 
 | | The structural and thermodynamic basis for the binding of TMC114, a next-generation HIV-1 protease inhibitor. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | King, N.M, Prabu-Jeyabalan, M, Nalivaika, E.A, Wigerinck, P.B.T.P, De Bethune, M.-P, Schiffer, C.A. | | Deposit date: | 2004-05-10 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor
J.Med.Chem., 48, 2005
|
|
1ITO
 
 | | Crystal Structure Analysis of Bovine Spleen Cathepsin B-E64c complex | | Descriptor: | Cathepsin B, N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-2-METHYL-BUTANE | | Authors: | Yamamoto, A, Tomoo, T, Matsugi, K, Hara, T, In, Y, Murata, M, Kitamura, K, Ishida, T. | | Deposit date: | 2002-01-19 | | Release date: | 2003-01-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | Structural basis for development of cathepsin B-specific noncovalent-type inhibitor: crystal structure of cathepsin B-E64c complex
BIOCHIM.BIOPHYS.ACTA, 1597, 2002
|
|
3W13
 
 | | Insulin receptor ectodomain construct comprising domains L1-CR in complex with high-affinity insulin analogue [D-PRO-B26]-DTI-NH2, alphact peptide(693-719) and FAB 83-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Lawrence, M.C, Smith, B.J, Brzozowski, A.M. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.303 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor
Nature, 493, 2013
|
|
4Q46
 
 | |
3W12
 
 | | Insulin receptor ectodomain construct comprising domains L1-CR in complex with high-affinity insulin analogue [D-PRO-B26]-DTI-NH2, alpha-CT peptide(704-719) and FAB 83-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | Authors: | Lawrence, M.C, Smith, B.J, Brzozowsk, A.M. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.301 Å) | | Cite: | How insulin engages its primary binding site on the insulin receptor
Nature, 493, 2013
|
|
1NG5
 
 | |
1OGB
 
 | | Chitinase b from Serratia marcescens mutant D142N | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Rao, F.V, Peter, M.G, Synstad, B, van Aalten, D.M.F, Eijsink, V.G.H. | | Deposit date: | 2003-04-29 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the D142N Mutant of the Family 18 Chitinase Chib from Serratia Marcescens and its Complex with Allosamidin
Biochim.Biophys.Acta, 1696, 2004
|
|
2BFZ
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20mM ZnSO4 in buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | AZIDE ION, BETA-LACTAMASE II, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-16 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
3AQC
 
 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium and FPP analogue | | Descriptor: | (2E,6E)-7,11-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, ... | | Authors: | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
2X5V
 
 | | 80 microsecond laue diffraction snapshot from crystals of a photosynthetic reaction centre 3 millisecond following photoactivation. | | Descriptor: | BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, FE (II) ION, ... | | Authors: | Wohri, A.B, Katona, G, Johansson, L.C, Fritz, E, Malmerberg, E, Andersson, M, Vincent, J, Eklund, M, Cammarata, M, Wulff, M, Davidsson, J, Groenhof, G, Neutze, R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Light-Induced Structural Changes in a Photosynthetic Reaction Center Caught by Laue Diffraction.
Science, 328, 2010
|
|
7WLC
 
 | |
