7FIS
 
 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with mannose 1-phosphate (M1P) | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, Beta-1,2-mannobiose phosphorylase, GLYCEROL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
2AZQ
 
 | | Crystal Structure of Catechol 1,2-Dioxygenase from Pseudomonas arvilla C-1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, FE (III) ION, catechol 1,2-dioxygenase | | Authors: | Earhart, C.A, Vetting, M.W, Gosu, R, Michaud-Soret, I, Que, L, Ohlendorf, D.H. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of catechol 1,2-dioxygenase from Pseudomonas arvilla
Biochem.Biophys.Res.Commun., 338, 2005
|
|
5B72
 
 | | Crystal structure of bovine lactoperoxidase with a broken covalent bond between Glu258 and heme moiety at 1.98 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution
Biochim. Biophys. Acta, 1865, 2016
|
|
6C9E
 
 | |
3INL
 
 | | Human Mitochondrial Aldehyde Dehydrogenase Asian Variant, ALDH2*2, complexed with agonist Alda-1 | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase, mitochondrial, ... | | Authors: | Perez-Miller, S, Hurley, T.D. | | Deposit date: | 2009-08-12 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3INJ
 
 | |
5UCP
 
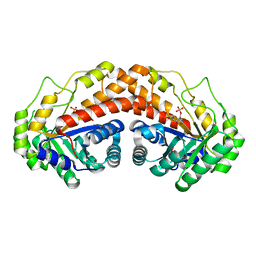 | | Class II fructose-1,6-bisphosphate aldolase E142A variant of Helicobacter pylori with FBP and cleavage products | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, 1,6-di-O-phosphono-D-fructose, Fructose-bisphosphate aldolase, ... | | Authors: | Jacques, B, Sygusch, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
5UCN
 
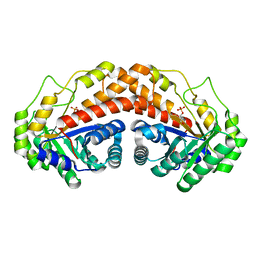 | | Class II fructose-1,6-bisphosphate aldolase E142A variant of Helicobacter pylori with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Jacques, B, Sygusch, J. | | Deposit date: | 2016-12-22 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
3G5N
 
 | | Triple ligand occupancy crystal structure of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole | | Descriptor: | 1-(biphenyl-4-ylmethyl)-1H-imidazole, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Sun, L, Maekawa, K, Halpert, J.R, Stout, C.D. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: ligand-induced structural response through alpha-helical repositioning.
Biochemistry, 48, 2009
|
|
3G93
 
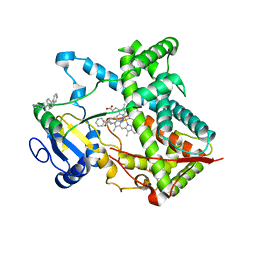 | | Single ligand occupancy crystal structure of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole | | Descriptor: | 1-(biphenyl-4-ylmethyl)-1H-imidazole, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Sun, L, Maekawa, K, Halpert, J.R, Stout, C.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: ligand-induced structural response through alpha-helical repositioning.
Biochemistry, 48, 2009
|
|
5UD3
 
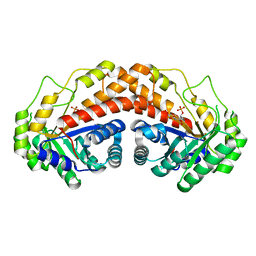 | | Class II fructose-1,6-bisphosphate aldolase H180Q variant of Helicobacter pylori with FBP | | Descriptor: | 1,6-di-O-phosphono-D-fructose, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Jacques, B, Sygusch, J. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
2WD8
 
 | | PTERIDINE REDUCTASE 1 (PTR1) FROM TRYPANOSOMA BRUCEI IN COMPLEX WITH NADP AND DDD00071204 | | Descriptor: | 1-(3,4-DICHLOROBENZYL)-7-PHENYL-1H-BENZIMIDAZOL-2-AMINE, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Robinson, D.A, Wyatt, P.G, Spinks, D, Brenk, R. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | One Scaffold, Three Binding Modes: Novel and Selective Pteridine Reductase 1 Inhibitors Derived from Fragment Hits Discovered by Virtual Screening.
J.Med.Chem., 52, 2009
|
|
4ROX
 
 | |
3IJ3
 
 | | 1.8 Angstrom Resolution Crystal Structure of Cytosol Aminopeptidase from Coxiella burnetii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytosol aminopeptidase, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Cytosol Aminopeptidase from Coxiella burnetii
TO BE PUBLISHED
|
|
3K4V
 
 | | New crystal form of HIV-1 Protease/Saquinavir structure reveals carbamylation of N-terminal proline | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Olajuyigbe, F.M, Demitri, N, Ajele, J.O, Maurizio, E, Randaccio, L, Geremia, S. | | Deposit date: | 2009-10-06 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Carbamylation of N-terminal proline.
ACS Med Chem Lett, 1, 2010
|
|
5UD4
 
 | | Class II fructose-1,6-bisphosphate aldolase H180Q variant of Helicobacter pylori with TBP | | Descriptor: | 1,6-di-O-phosphono-D-tagatose, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Jacques, B, Sygusch, J. | | Deposit date: | 2016-12-23 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
1P1J
 
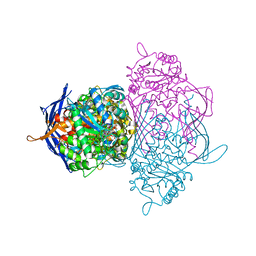 | | Crystal structure of the 1L-myo-inositol 1-phosphate synthase complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Inositol-3-phosphate synthase, ... | | Authors: | Jin, X, Geiger, J.H. | | Deposit date: | 2003-04-12 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of NAD(+)- and NADH-bound 1-l-myo-inositol 1-phosphate synthase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4I42
 
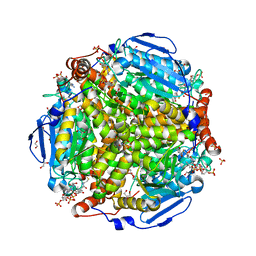 | | E.coli. 1,4-dihydroxy-2-naphthoyl coenzyme A synthase (ecMenB) in complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, 1-hydroxy-2-naphthoyl-CoA, ... | | Authors: | Sun, Y, Song, H, Li, J, Li, Y, Jiang, M, Zhou, J, Guo, Z. | | Deposit date: | 2012-11-27 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme A synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4K26
 
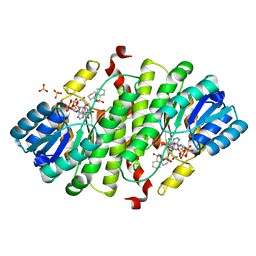 | | 4,4-Dioxo-5,6-dihydro-[1,4,3]oxathiazines, a novel class of 11 -HSD1 inhibitors for the treatment of diabetes | | Descriptor: | (4aS,8aR)-N-cyclohexyl-4a,5,6,7,8,8a-hexahydro-4,1,2-benzoxathiazin-3-amine 1,1-dioxide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Loenze, P, Schimanski-Breves, S, Vonderheyden, C, Engel, C.K. | | Deposit date: | 2013-04-08 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 1,1-Dioxo-5,6-dihydro-[4,1,2]oxathiazines, a novel class of 11-HSD1 inhibitors for the treatment of diabetes.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4K1L
 
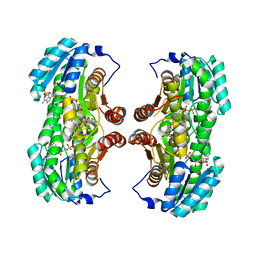 | | 4,4-Dioxo-5,6-dihydro-[1,4,3]oxathiazines, a novel class of 11 beta-HSD1 inhibitors for the treatment of diabetes | | Descriptor: | (4aS,8aR)-N-cyclohexyl-4a,5,6,7,8,8a-hexahydro-4,1,2-benzoxathiazin-3-amine 1,1-dioxide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Loenze, P, Schimanski-Breves, S, Von der Heyden, C, Engel, C.K. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 1,1-Dioxo-5,6-dihydro-[4,1,2]oxathiazines, a novel class of 11-HSD1 inhibitors for the treatment of diabetes.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5PGV
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE | | Descriptor: | 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
4QUS
 
 | | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase YpeA, PHOSPHATE ION | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Flores, K.J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655
To be Published
|
|
1P1K
 
 | |
5OFM
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-amino-1-methyl-1H-indole | | Descriptor: | 1-methylindol-5-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-amino-1-methyl-1H-indole
To be published
|
|
7KH6
 
 | |
