7K0C
 
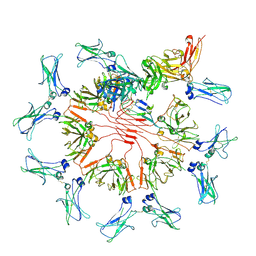 | | Structure of Secretory IgM Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, Immunoglobulin heavy constant mu, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-20 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human secretory immunoglobulin M core.
Structure, 29, 2021
|
|
1LY4
 
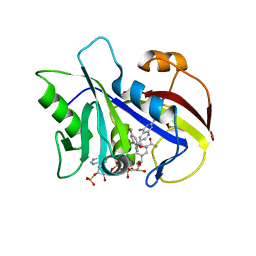 | | Analysis of quinazoline and PYRIDO[2,3D]PYRIMIDINE N9-C10 reversed bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1M1U
 
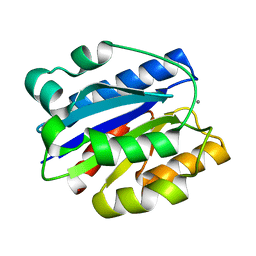 | | AN ISOLEUCINE-BASED ALLOSTERIC SWITCH CONTROLS AFFINITY AND SHAPE SHIFTING IN INTEGRIN CD11B A-DOMAIN | | Descriptor: | CALCIUM ION, Integrin alpha-M | | Authors: | Xiong, J.-P, Li, R, Essafi, M, Stehle, T, Arnaout, M.A. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An isoleucine-based allosteric switch controls affinity and shape shifting in integrin CD11b A-domain.
J.Biol.Chem., 275, 2000
|
|
8VC5
 
 | |
2IMN
 
 | |
2IMM
 
 | |
4WR7
 
 | | Crystal structure of human carbonic anhydrase isozyme I with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide. | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide, ACETATE ION, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-10-23 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
4WUQ
 
 | | Crystal structure of human carbonic anhydrase isozyme I with 2,3,5,6-Tetrafluoro-4-piperidin-1-ylbenzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-(piperidin-1-yl)benzenesulfonamide, Carbonic anhydrase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
3RN6
 
 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with zinc and isoguanine | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, 6-amino-3,7-dihydro-2H-purin-2-one, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Rescue of the orphan enzyme isoguanine deaminase.
Biochemistry, 50, 2011
|
|
4LNW
 
 | | Crystal structure of TR-alpha bound to T3 in a second site | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor alpha | | Authors: | Puhl, A.C, Aparicio, R, Polikarpov, I. | | Deposit date: | 2013-07-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a new hormone-binding site on the surface of thyroid hormone receptor.
Mol.Endocrinol., 28, 2014
|
|
4LNX
 
 | | Crystal structure of TR-alpha bound to T4 in a second site | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor alpha | | Authors: | Puhl, A.C, Aparicio, R, Polikarpov, I. | | Deposit date: | 2013-07-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identification of a new hormone-binding site on the surface of thyroid hormone receptor.
Mol.Endocrinol., 28, 2014
|
|
2QCQ
 
 | |
4W4Z
 
 | | Structure of the EphA4 LBD in complex with peptide | | Descriptor: | APY-bAla8.am peptide, Ephrin type-A receptor 4, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Mace, P.D, Riedl, S.J. | | Deposit date: | 2014-08-15 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Development and Structural Analysis of a Nanomolar Cyclic Peptide Antagonist for the EphA4 Receptor.
Acs Chem.Biol., 9, 2014
|
|
6UL3
 
 | |
6UKC
 
 | |
6DRC
 
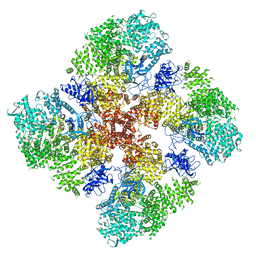 | | High IP3 Ca2+ human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DQS
 
 | | Class 3 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3HY4
 
 | | Structure of human MTHFS with N5-iminium phosphate | | Descriptor: | 5-formyltetrahydrofolate cyclo-ligase, MAGNESIUM ION, N-({trans-4-[({(2R,4R,4aS,6S,8aS)-2-amino-4-hydroxy-5-[(phosphonooxy)methyl]decahydropteridin-6-yl}methyl)amino]cyclohexyl}carbonyl)-L-glutamic acid, ... | | Authors: | Wu, D, Li, Y, Song, G, Cheng, C, Shaw, N, Liu, Z.-J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Structural basis for the inhibition of human 5,10-methenyltetrahydrofolate synthetase by N10-substituted folate analogues
Cancer Res., 69, 2009
|
|
8J0P
 
 | | Chitin binding SusD-like protein AqSusD from a marine Bacteroidetes | | Descriptor: | Chitin binding SusD-like protein | | Authors: | Yang, J. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
7QX9
 
 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 65 | | Descriptor: | 3-[[4-(4-ethanoyl-3,5-dimethyl-1~{H}-pyrrol-2-yl)-1,3-thiazol-2-yl]methyl]imidazolidine-2,4-dione, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
2QEB
 
 | | Crystal Structure of Anopheles Gambiae D7R4-Histamine Complex | | Descriptor: | D7R4 Protein, GLYCEROL, HISTAMINE | | Authors: | Andersen, J.F, Mans, B.J, Calvo, E, Ribeiro, J.M. | | Deposit date: | 2007-06-25 | | Release date: | 2007-10-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The Crystal Structure of D7r4, a Salivary Biogenic Amine-binding Protein from the Malaria Mosquito Anopheles gambiae
J.Biol.Chem., 282, 2007
|
|
6UGL
 
 | | VqmA bound to DPO | | Descriptor: | 3,5-dimethylpyrazin-2(1H)-one, Helix-turn-helix transcriptional regulator | | Authors: | Paczkowski, J.E, Huang, X. | | Deposit date: | 2019-09-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism underlying autoinducer recognition in theVibrio choleraeDPO-VqmA quorum-sensing pathway.
J.Biol.Chem., 295, 2020
|
|
2QEV
 
 | | Crystal Structure of Anopheles gambiae D7r4 | | Descriptor: | D7R4 Protein | | Authors: | Andersen, J.F, Mans, B.J, Calvo, E, Ribeiro, J.M. | | Deposit date: | 2007-06-26 | | Release date: | 2007-10-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | The Crystal Structure of D7r4, a Salivary Biogenic Amine-binding Protein from the Malaria Mosquito Anopheles gambiae
J.Biol.Chem., 282, 2007
|
|
6KSP
 
 | |
1MC5
 
 | | Ternary complex of Human glutathione-dependent formaldehyde dehydrogenase with S-(hydroxymethyl)glutathione and NADH | | Descriptor: | 2-AMINO-4-[1-CARBOXYMETHYL-CARBAMOYL)-2-HYDROXYMETHYLSULFANYL-ETHYLCARBAMOYL]-BUTYRIC ACID, Alcohol dehydrogenase class III chi chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sanghani, P.C, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-08-05 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase.
Structural changes associated with Ternary Complex formation
Biochemistry, 41, 2002
|
|
