8EUB
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2 and GDP, Structure I | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-18 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EVQ
 
 | | Hypopseudouridylated Ribosome bound with TSV IRES, eEF2, GDP, and sordarin, Structure I | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EVR
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2, GDP, and sordarin, Structure II | | Descriptor: | (1S,3S,3aR,4S,4aR,7R,7aR,8aS)-8a-{[(6-deoxy-4-O-methyl-alpha-D-altropyranosyl)oxy]methyl}-4-formyl-7-methyl-3-(propan-2-yl)decahydro-1,4-methano-s-indacene-3a(1H)-carboxylate, 18S rRNA, 25S rRNA, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EVS
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2 and GDP, Structure II | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EWB
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), eEF2 and GDP, Structure III | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EWC
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES), Structure II | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8EVT
 
 | | Hypopseudouridylated yeast 80S bound with Taura syndrome virus (TSV) internal ribosome entry site (IRES) refined against a composite map | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Zhao, Y, Rai, J, Li, H. | | Deposit date: | 2022-10-20 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Regulation of translation by ribosomal RNA pseudouridylation.
Sci Adv, 9, 2023
|
|
8FY8
 
 | | 5-MeO-DMT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | 2-(5-methoxy-1H-indol-3-yl)-N,N-dimethylethan-1-amine, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-25 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
7TD0
 
 | | Lysophosphatidic acid receptor 1-Gi complex bound to LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7T6S
 
 | | Structure of the human FPR2-Gi complex with compound C43 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6V
 
 | | Structure of the human FPR2-Gi complex with fMLFII | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6U
 
 | | Structure of the human FPR2-Gi complex with CGEN-855A | | Descriptor: | B9-scFv, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6T
 
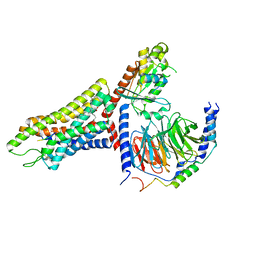 | | Structure of the human FPR1-Gi complex with fMLFII | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6B
 
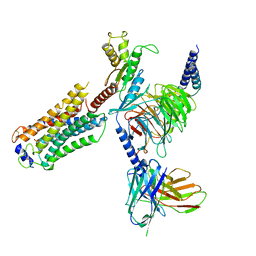 | | Structure of S1PR2-heterotrimeric G13 signaling complex | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, X, Chen, H. | | Deposit date: | 2021-12-13 | | Release date: | 2022-04-06 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of S1PR2-heterotrimeric G 13 signaling complex.
Sci Adv, 8, 2022
|
|
7SQO
 
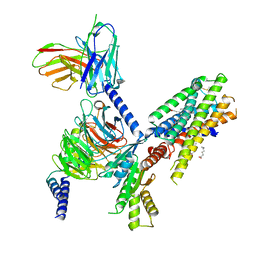 | | Structure of the orexin-2 receptor(OX2R) bound to TAK-925, Gi and scFv16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | McGrath, A.P, Kang, Y, Flinspach, M. | | Deposit date: | 2021-11-05 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
7SR8
 
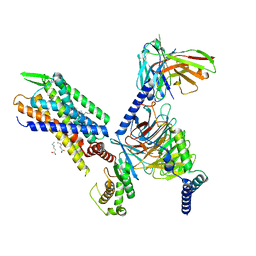 | | Molecular mechanism of the the wake-promoting agent TAK-925 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hypocretin receptor type 2, ... | | Authors: | Yin, J, Chapman, K, Lian, P, De Brabander, J.K, Rosenbaum, D.M. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
7SUK
 
 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | Descriptor: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-07-06 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYJ
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 4(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYX
 
 | | Structure of the delta dII IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S24, 40S ribosomal protein S25, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYL
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, closed conformation. Structure 6(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SRR
 
 | | 5-HT2B receptor bound to LSD in complex with heterotrimeric mini-Gq protein obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2B, G protein subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Barros-Alvarez, X, Kim, K, Panova, O, Cao, C, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-11-08 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD.
Neuron, 110, 2022
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYW
 
 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYQ
 
 | | Structure of the wt IRES and 40S ribosome ternary complex, open conformation. Structure 11(wt) | | Descriptor: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
