6HL1
 
 | | Crystal Structure of Farnesoid X receptor (FXR) with bound NCoA-2 peptide and CDCA | | Descriptor: | Bile acid receptor, CHENODEOXYCHOLIC ACID, NCoA-2 peptide (Nuclear receptor coactivator 2), ... | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Molecular tuning of farnesoid X receptor partial agonism.
Nat Commun, 10, 2019
|
|
6SUO
 
 | | ERa_L536S (L536S/C381S/C471S,C530S) in complex with a tricyclic indole (compound 6) | | Descriptor: | (~{E})-3-[3,5-bis(fluoranyl)-4-[(1~{R},3~{R})-2-(2-fluoranyl-2-methyl-propyl)-1,3-dimethyl-4,9-dihydro-3~{H}-pyrido[3,4-b]indol-1-yl]phenyl]prop-2-enoic acid, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Building Bridges in a Series of Estrogen Receptor Degraders: An Application of Metathesis in Medicinal Chemistry.
Acs Med.Chem.Lett., 10, 2019
|
|
7LUI
 
 | |
7LSM
 
 | | Crystal structure of E.coli DsbA in complex with bile salt taurocholate | | Descriptor: | DI(HYDROXYETHYL)ETHER, TAUROCHOLIC ACID, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Selective Binding of Small Molecules to Vibrio cholerae DsbA Offers a Starting Point for the Design of Novel Antibacterials.
Chemmedchem, 17, 2022
|
|
6HVO
 
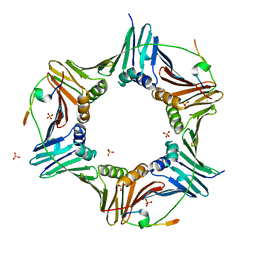 | | Crystal structure of human PCNA in complex with three peptides of p12 subunit of human polymerase delta | | Descriptor: | DNA polymerase delta subunit 4, Proliferating cell nuclear antigen, SULFATE ION | | Authors: | Gonzalez-Magana, A, Romano-Moreno, M, Rojas, A.L, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-23 | | Last modified: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The p12 subunit of human polymerase delta uses an atypical PIP box for molecular recognition of proliferating cell nuclear antigen (PCNA).
J.Biol.Chem., 294, 2019
|
|
6TH7
 
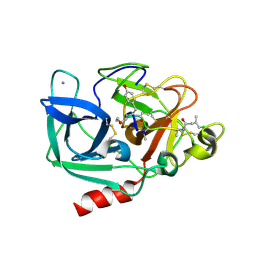 | |
6I6O
 
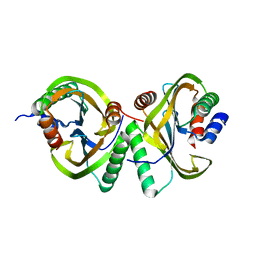 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I6Y
 
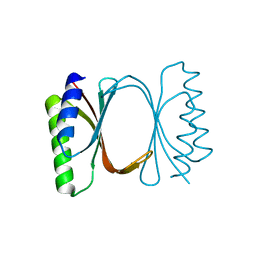 | |
7LC0
 
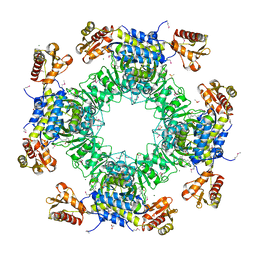 | | Structure of D-Glucosaminate-6-phosphate Ammonia-lyase | | Descriptor: | ACETIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2021-01-09 | | Release date: | 2021-06-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Mechanism of d-Glucosaminate-6-phosphate Ammonia-lyase: A Novel Octameric Assembly for a Pyridoxal 5'-Phosphate-Dependent Enzyme, and Unprecedented Stereochemical Inversion in the Elimination Reaction of a d-Amino Acid.
Biochemistry, 60, 2021
|
|
7LCE
 
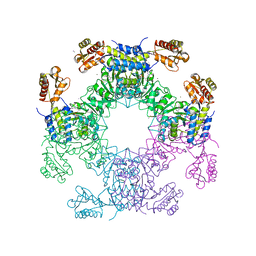 | | Structure of D-Glucosaminate-6-phosphate Ammonia-lyase | | Descriptor: | CALCIUM ION, D-glucosaminate-6-phosphate ammonia lyase, DIMETHYL SULFOXIDE | | Authors: | Phillips, R.S. | | Deposit date: | 2021-01-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure and Mechanism of d-Glucosaminate-6-phosphate Ammonia-lyase: A Novel Octameric Assembly for a Pyridoxal 5'-Phosphate-Dependent Enzyme, and Unprecedented Stereochemical Inversion in the Elimination Reaction of a d-Amino Acid.
Biochemistry, 60, 2021
|
|
6U11
 
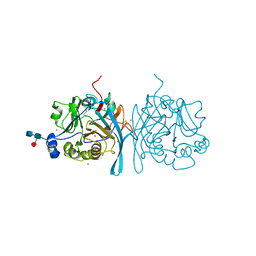 | | Xenopus laevis N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (NAGPA) (C46S C219S C453S C480S C486S) with CTD mostly flexible | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Mannose-6-Phosphate Uncovering Enzyme.
Structure, 28, 2020
|
|
6I6W
 
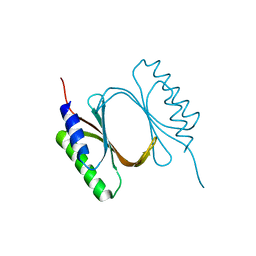 | |
6SLO
 
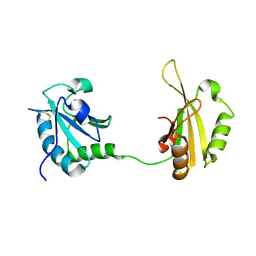 | | Crystal structure of PUF60 UHM domain in complex with 7,8 dimethoxyperphenazine | | Descriptor: | 2-[4-[3-(8-chloranyl-2,3-dimethoxy-phenothiazin-10-yl)propyl]piperazin-1-yl]ethanol, MAGNESIUM ION, Thioredoxin,Poly(U)-binding-splicing factor PUF60 | | Authors: | Jagtap, P.K.A, Kubelka, T, Bach, T, Sattler, M. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of phenothiazine derivatives as UHM-binding inhibitors of early spliceosome assembly.
Nat Commun, 11, 2020
|
|
6RMC
 
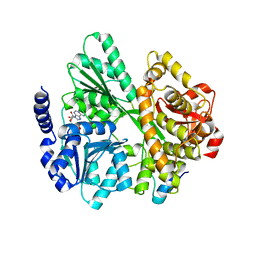 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative mRNA splicing factor, ... | | Authors: | Hamann, F, Neumann, P, Schmitt, A, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UIK
 
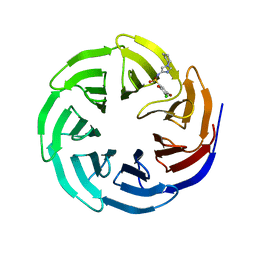 | |
6UIF
 
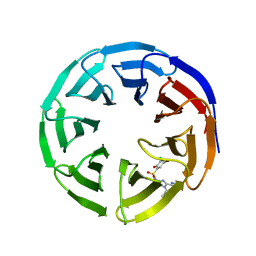 | |
7MRU
 
 | | Crystal structure of S62A MIF2 mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-dopachrome decarboxylase | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MW7
 
 | | Crystal structure of P1G mutant of D-dopachrome tautomerase | | Descriptor: | D-dopachrome decarboxylase, SODIUM ION, SULFATE ION | | Authors: | Manjula, R, Murphy, E.L, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-15 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MSE
 
 | | High-resolution crystal structure of hMIF2 with tartrate at the active site | | Descriptor: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MRV
 
 | | F100A mutant structure of MIF2 (D-DT) | | Descriptor: | D-dopachrome decarboxylase, SULFATE ION | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7L3R
 
 | |
7M98
 
 | | ATAD2 bromodomain complexed with histone H4K5ac (res 1-10) ligand | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4 | | Authors: | Malone, K.L, Phillips, M, Nix, J.C, Glass, K.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coordination of Di-Acetylated Histone Ligands by the ATAD2 Bromodomain.
Int J Mol Sci, 22, 2021
|
|
6RM8
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TYY
 
 | | Hedgehog autoprocessing mutant D46H | | Descriptor: | Protein hedgehog | | Authors: | Li, H, Li, Z, Wang, C, Callahan, B.P. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | General Base Swap Preserves Activity and Expands Substrate Tolerance in Hedgehog Autoprocessing.
J.Am.Chem.Soc., 141, 2019
|
|
6UJH
 
 | |
